
Lung cancer A549 cells suppressed with overexpressed HNF1B or PCDHA13 inhibited PI3K/AKT phosphorylation
Introduction
Lung cancer is one of the most serious types of cancer and is responsible for one-fifth of all cancer-related mortality (1). In recent years, the lack of suitable and reliable biomarkers and therapeutic targets, delays in diagnosis, and ineffectiveness of drugs have become obstacles to the progress of lung cancer treatment (2). In addition, the development of cancer resistance has made treating lung cancer patients more challenging (3). Currently, the most common treatments for lung cancer are surgical resection, radiotherapy, and chemotherapy, although their curative effect is unsatisfactory, and lung cancer patients can expect a 5-year overall survival rate of only 15% (4). Therefore, it is crucial that a new, more effective treatment for lung cancer is discovered.
As an important transcription factor in the regulation of embryonic survival and vertebrate development, hepatocyte nuclear factor 1B (HNF1B) is reported to be closely associated with the risk of many cancers, including prostate (5), ovarian (6,7), endometrial (8,9), and lung cancer (10). The overexpression of HNF1B has recently been demonstrated to be a potential contributing factor to the prevention of prostate cancer (11). PCDHA13 is a member of the protocadherin (PCDH) family involved in cell adhesion and signal transduction. The abnormal hypermethylation of PCDHA13 promoter has been detected in a variety of tumor tissues and cells (12). Some non-clustered PCHD genes have been identified as tumor suppressor genes, and their abnormal methylation is closely related to tumor occurrence and development (13,14). Both HNF1B and PCDH were implicated in PI3K/AKT signaling. For example, the epidermal growth factor (EGF) and AKT mediated nuclear translocation of zyxin activated HNF-1B-dependent transcription (15); and PCDH10 inhibits hepatocellular carcinoma cell proliferation and induces cell apoptosis by inhibiting the PI3K/Akt signaling pathway (16). However, the role of HNF1B and PCDH across tumors seemed varied. The relationship between HNF1B and PCDH and lung cancer has not yet been established. Therefore, this study investigated the effects of HNF1B and PCDHA13 overexpression on the phosphorylation of PI3K/AKT and malignant biological behavior in lung cancer A549 cells, with the aim of providing an experimental basis on which to explore a new, more effective treatment for patients with lung cancer.
Methods
Reagents and instruments
A549 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, Manassas). Dulbecco’s Modified Eagle’s medium (DMEM), fetal calf serum, and pancreatin were purchased from Invitrogen (Invitrogen, Waltham, MA, USA). Bicinchoninic acid (BCA) protein quantification kit was purchased from Pierce (PIERCE, Illinois, USA). MatrigelTM basement membrane matrix, Transwell chamber and Matrigel and FACS Vantage SE flow cytometer were purchased from BD Biosciences (BD Biosciences, NJ, USA). Antibodies for Survivin, PCNA, Caspase3, Caspase9, PI3K, p-PI3K, AKT and p-AKT were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). HRP-labeled goat anti-rat secondary antibody was purchased from Santa Cruz (Santa Cruz Biotechnology Dallas, TX, USA). Apoptosis detection kit (Annexin PE/7-AAD) was purchased from Kaiji (Nanjing Kaiji Biological Co., Ltd., China). High speed refrigerated centrifuge was from Beckman (Beckman, Coulter, CA, USA). CO2 incubator was from Thermo (Thermo Fisher Scientific, Waltham, MA, USA). Electrophoresis chamber and Trans-Blot Turbo were purchased from Bio-Rad (Bio-Rad, Hercules, CA, USA). Gel imaging system GDS-800 was purchased from UVP (1UVP, LLC, Upland, CA, USA).
Cell culture
A549 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum. All of the cells were cultured at 37 °C in a humidified incubator containing 5% CO2. When the cells reached over 85% confluence, they were digested with 0.25% trypsin for passaging.
RT-PCR
HNF1B and PCDHA13 were amplified using designed primers listed in Table 1. Then the amplified RNAs were used to construct pcDNA overexpression plasmids, which were divided into four groups: the control group, the pcNDA group, the pcNDA-HNF1B group, and the pcNDA-PCDHA13 group. Then the PCR amplified products were cut and collected, added with pcDNA3.1, Apa I and Nhe I, and then digested and ligated. After transfected with the products, DH5α competent cells were cultured with the ampicillin agar plates at 37 °C. After expanded culture, cells were harvested for extraction of recombinant plasmid. HNF1B and PCDHA13 mRNA levels were detected using RT-PCR, to confirm that the overexpression was successful. For RT-PCR, Trizol reagent was used to extract total RNA, and the absorbance was measured at 260 and 280 nm with an ultraviolet spectrophotometer. cDNA was synthesized using the Super RT cDNA Kit, in line with the manufacturer’s instructions. Quantitative real-time PCRs were also performed, using an ABI 7500 system with the Quantifast® SYBR® Green PCR Kit at 95 °C for 15 s and 60 °C for 60 s.
Table 1
| Name | Primer sequence |
|---|---|
| HNF1B | Forward: 5'-CCCCTCACCATCAGCCAAG-3' |
| Reverse: 5'-GGTTCTGAGATTGCTGGGGATT-3' | |
| PCDHA13 | Forward: 5'-GAGGAGGACTCAGAATGCTTG-3' |
| Reverse: 5'-GGGTTGCCTGGTCCGTAT-3' | |
| GAPDH | Forward: 5'-AGGTCGGTGTGAACGGATTTG-3' |
| Reverse: 5'-GGGGTCGTTGATGGCAACA-3' |
Clone formation experiments
A549 cells were seeded into 6-well plates at a density of 1,000 cells/well and incubated for 10 days. The medium was changed every 3 days throughout the cultivation process. On the last day, the cell colonies were washed twice, fixed with 3 mL of methanol for 15 min, and 1 mL of 0.1% crystal violet was added to each well for 15 min. After washing 3 times with PBS, the cell clones were observed and counted under a microscope.
Flow cytometry
The apoptosis rate in A549 cells was measured and analyzed with Annexin V-FITC apoptosis detection kit. Cells were cultured in a 6-well plate at a maximum density of 3×106 cells/well for 48 h, with different concentrations of dihydromyricetin or dimethyl sulfoxide as a control. The cells were then incubated with hydrogen peroxide to induce oxidative stress at 37 °C with 5% CO2. Next, the cells were washed 3 times with cold PBS and treated with 100 µL of binding buffer, before incubation with 3 µL Annexin V-FITC and 10 µL propidium iodide for 20 minutes. Flow cytometry was performed to detect apoptosis.
Transwell assay
The cells in each group were digested, centrifuged, and resuspended in serum-free DMEM medium. Then 1×104 cells were seeded into the upper chamber of each well. The medium with serum was added to the lower chamber. After incubation for 48 h, the cells in the upper chamber were wiped with a sterile cotton swab. The Transwell chamber was air-dried upside down and placed into a 12-well plate with 500 µL of staining solution (containing 0.1% crystal violet) for 20 min. The chamber was removed from the staining buffer, before being washed with PBS 3 times and air-dried. Five fields of view were taken around the diameter of the Transwell chamber and photographed under a microscope to measure cell migration.
Western blotting
Cell samples were lysed using protein lysis buffer. Then, the total protein was measured using the BCA protein assay kit (Beyotime). A sample containing 20 mg of protein was separated running 10% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. After transfection, the membranes were blocked with 5% skim milk at room temperature for 2 h, and then incubated with primary antibodies overnight at 4 °C. The primary antibodies and dilutions were: Survivin, 1:1,000; PCNA, 1:1,000; Caspase3, 1:1,000; Caspase9, 1:1,000; PI3K, 1:1,000; p-PI3K 1:1,000; AKT, 1:1,000; and p-AKT, 1:1,000. The secondary antibody was added at room temperature for 1 h the following day. Finally, ECL reagent was added to detect the intensity of chemiluminescence of the target bands with set exposure parameters.
Statistical methods
SPSS 19.0 was used to perform statistical analysis of all experimental data. Differences between groups were tested using t-test or one-way analysis of variance. The data were processed as mean ± standard deviation (), and P<0.05 was considered statistically significant.
Results
Changes in HNF1B and PCDHA13 expression in lung cancer A549 cells
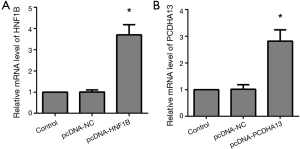
The mRNA level of HNF1B in the pcNDA-HNF1B group was significantly higher than that in the control group (P<0.05), and the mRNA level of PCDHA13 in the pcNDA-PCDHA13 group was significantly higher than that in the control group (P<0.05; Figure 1).
The effect of HNF1B and PCDHA13 overexpression on proliferation of lung cancer A549 cells
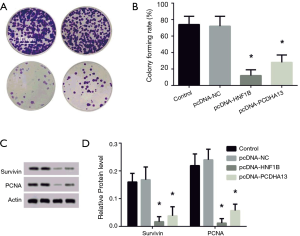
The colony formation rates of the pcNDA-HNF1B group and the pcNDA-PCDHA13 group were significantly reduced in comparison with that of the control group (P<0.05; Figure 2A). The levels of survivin and PCNA expression in the PCNDA-HNF1B and pcNDA-PCDHA13 groups were significantly lower than that of the control group (P<0.05; Figure 2B).
The effect of HNF1B and PCDHA13 overexpression on apoptosis of lung cancer A549 cells
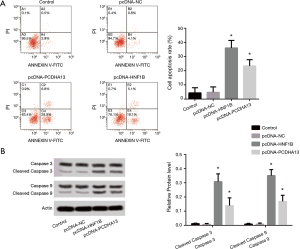
The apoptosis rates of the pcNDA-HNF1B group and the pcNDA-PCDHA13 group were significantly increased compared with that of the control group (P<0.05; Figure 3A). In the pcNDA-HNF1B and pcNDA-PCDHA13 groups, the ratios of cleaved caspase3/caspase3 and cleaved caspase9/caspase9 protein expression levels were significantly increased compared to that in the control group (P<0.05; Figure 3B).
The effect of HNF1B and PCDHA13 overexpression on invasion of lung cancer A549 cells
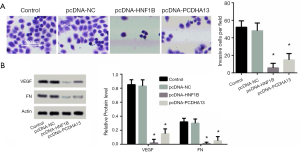
In comparison to the control group, cell invasion in the pcNDA-HNF1B group and the pcNDA-PCDHA13 group was significantly reduced (P<0.05; Figure 4A), as indicated with Transwell. The reduced invasive cells with HNF1B and PCDHA13 overexpression were further validated with the expression levels of VEGF and fibronectin protein (P<0.05; Figure 4B).
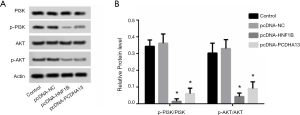
The effect of HNF1B and PCDHA13 overexpression on the phosphorylation of PI3K/AKT
The phosphorylation levels of PI3K and AKT in the pcNDA-HNF1B and pcNDA-PCDHA13 groups were significantly lower than those in the control group (P<0.05; Figure 5).
Discussion
HNF1B, a member of the transcription factor superfamily, contains the homeodomain, which is involved in regulating the expression of various genes in a tissue-specific manner during the development of different organs and embryos (17). The gene mapping database reports that HNF1B mRNA is highly expressed in the kidney and islets (17). Studies have investigated the relationship between HNF1B locus (located on chromosome 17q12) and the risk of cancer (8). PCDHA13 is involved in cell adhesion and signaling transduction, and the abnormal hypermethylation of its promoter has been detected in a range of different tumor tissues and cells (12). However, the relationship between clustered PCDH families and tumors is not yet fully understood. In the present study, the expression levels of HNF1B and PCDHA13 observed in A549 cells indicated that HNF1B and PCDHA13 overexpression is potentially related to the malignant biological behavior of lung cancer cells.
The infinite proliferation of cancer cells is an important element in cancer development. Survivin is a regulatory gene that is highly expressed during cell mitosis, which can promote the normal progress of cell mitosis and play a role in the promotion of cell proliferation. Currently, PCNA is recognized as a cell proliferation marker protein, which is highly expressed during cell proliferation. The downregulation of survivin and PCNA has been reported to be a hallmark of decreased cell proliferation (18,19). Studies have found that with as the tumor progresses, the initial tumor protection function of HNF1B disappears and HNF1B expression is downregulated, inhibiting the proliferation of prostate cancer cell (20). The results of this study showed that the overexpression of HNF1B and PCDHA13 resulted in a decrease in the clonal formation rate of A549 cells and the protein levels of Survivin and PCNA, suggesting that HNF1B and PCDHA13 overexpression can inhibit A549 cell proliferation.
Inducing apoptosis is one of the main approaches to inhibiting tumor growth. Bcl-2, an anti-apoptotic protein, can regulate apoptosis via the regulation of caspase-9 and caspase-3 dependent pathways. When Bax is upregulated and Bcl-2 is downregulated, cytochrome C release can be stimulated in mitochondria with caspase-9 activation, which can activate caspase-3, eventually leading to apoptosis (21). The results of this study showed that the overexpression of HNF1B and PCDHA13 produced an increase in the apoptosis rate, as well as in the protein expression ratios of cleaved caspase3/caspase3 and cleaved caspase9/caspase9 protein, suggesting that HNF1B and PCDHA13 overexpression can induce A549 cell apoptosis, while the mechanism behind HNF1B and PCDHA13-induced apoptosis in A549 cells calls for further study.
Malignant tumor cells have the ability to infiltrate and invade surrounding normal tissue. As a key regulator of angiogenesis, VEGF is synthesized and secreted in the majority of cancer cells, which plays an important role in tumor growth and metastasis (22). Fibronectin, an invasive extracellular matrix protein, is enriched in the tumor microenvironment (23). It is reported that the methylation of HNF1B promoter is associated with SNP genotypes of prostate cancer risk and the expression of HNF1B (11). Overexpression of HNF1B significantly inhibits EZH2-mediated overgrowth and EMT processes, such as the migration and invasion of prostate cancer cell lines (24). The results of this study indicated that the overexpression of HNF1B and PCDHA13 caused a decrease in the number of invading cells and the protein levels of VEGF and fibronectin, which suggests HNF1B and PCDHA13 overexpression has an inhibitive effect on A549 cell invasion.
The cascade of the PI3K signaling pathway serves a core role in the occurrence and development of lung cancer (25), regulating a variety of cellular processes related to tumor cell growth and progression, including cell proliferation, migration, apoptosis, and angiogenesis (26). AKT is an important target molecular of the PI3K signaling pathway and represents the endpoints of a variety of growth factors and cytokine-induced signal transduction (27). This study showed that the overexpression of HNF1B and PCDHA13 oversaw a decrease in the phosphorylation of PI3K and AKT, suggesting that a relationship between the malignant biological behavior of A549 cells and the PI3K/AKT signaling pathway may exist. However, further study to explore the mechanism of this phenomenon is required.
The possible role of HNF1B and PCDHA13 in vivo still needs to be carefully validated as there are some controversial studies. In Kras (G12D) mutant mice, HNF1B increased the sensitivity to adenoma formation in the lung (28). The expression of HNF1B even varied in a tumor model across time. In TRAMP(+) mice, the expression of HNF1B at 18 weeks is markedly increased compared to its expression at 12 weeks (20). The clustered PCDH family members, on the other hand, were also implicated in both tumor promoting and suppressive events (29,30). The effects of HNF1B and PCDHA13 in lung cancers in vivo and in clinic remain largely undiscovered.
In summary, the overexpression of HNF1B and PCDHA13 genes can inhibit the phosphorylation of PI3K/AKT and hinder the malignant biological behavior of lung cancer A549 cells, providing an experimental basis for improvements in lung cancer treatment.
Acknowledgments
Funding: This research was financially supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1727
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1727). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen Y, Huang Y, Ning H, et al. Clinic-pathologic features and gene fusion pattern of ALK and ROS1 in non-small cell lung cancer show association with household coal combustion. Transl Cancer Res 2019;8:2164-74. [Crossref]
- Chen J, Soudy H. Osimertinib in the treatment of leptomeningeal disease in T790M-negative, epidermal growth factor receptor-mutated non-small cell lung cancer: a case report. Chin Clin Oncol 2019;8:29. [Crossref] [PubMed]
- Chen C, Huang X, Peng M, et al. Multiple primary lung cancer: a rising challenge. J Thorac Dis 2019;11:S523-36. [Crossref] [PubMed]
- Tong Y, Qu Y, Li S, et al. Cumulative evidence for relationships between multiple variants of HNF1B and the risk of prostate and endometrial cancers. BMC Med Genet 2018;19:128. [Crossref] [PubMed]
- Burghaus S, Fasching PA, Haberle L, et al. Genetic risk factors for ovarian cancer and their role for endometriosis risk. Gynecol Oncol 2017;145:142-7. [Crossref] [PubMed]
- Shen H, Fridley BL, Song H, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun 2013;4:1628. [Crossref] [PubMed]
- Painter JN, O'Mara TA, Batra J, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet 2015;24:1478-92. [Crossref] [PubMed]
- Mandato VD, Farnetti E, Torricelli F, et al. HNF1B polymorphism influences the prognosis of endometrial cancer patients: a cohort study. BMC Cancer 2015;15:229. [Crossref] [PubMed]
- Sun JZ, Yang XX, Hu NY, et al. Genetic Variants in MMP9 and TCF2 Contribute to Susceptibility to Lung Cancer. Chin J Cancer Res 2011;23:183-7. [Crossref] [PubMed]
- Ross-Adams H, Ball S, Lawrenson K, et al. HNF1B variants associate with promoter methylation and regulate gene networks activated in prostate and ovarian cancer. Oncotarget 2016;7:74734-46. [Crossref] [PubMed]
- Aghili L, Foo J, DeGregori J, et al. Patterns of somatically acquired amplifications and deletions in apparently normal tissues of ovarian cancer patients. Cell Rep 2014;7:1310-9. [Crossref] [PubMed]
- Zhang P, Wang H, Wang J, et al. Association between protocadherin 8 promoter hypermethylation and the pathological status of prostate cancer. Oncol Lett 2017;14:1657-64. [Crossref] [PubMed]
- Deng QK, Lei YG, Lin YL, et al. Prognostic Value of Protocadherin10 (PCDH10) Methylation in Serum of Prostate Cancer Patients. Med Sci Monit 2016;22:516-21. [Crossref] [PubMed]
- Choi YH, McNally BT, Igarashi P. Zyxin regulates migration of renal epithelial cells through activation of hepatocyte nuclear factor-1β. Am J Physiol Renal Physiol 2013;305:F100-10. [Crossref] [PubMed]
- Ye M, Li J, Gong J. Pcdh10 gene inhibits cell proliferation and induces cell apoptosis by inhibiting the PI3K/AKT signaling pathway in hepatocellular carcinoma cells. Oncol Rep 2017;37:3167-74. [Crossref] [PubMed]
- Heliot C, Desgrange A, Buisson I, et al. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development 2013;140:873-85. [Crossref] [PubMed]
- Su Y, Sun B, Lin X, et al. Therapeutic strategy with artificially-designed i-lncRNA targeting multiple oncogenic microRNAs exhibits effective antitumor activity in diffuse large B-cell lymphoma. Oncotarget 2016;7:49143-55. [Crossref] [PubMed]
- Singhal J, Nagaprashantha L, Chikara S, et al. 2'-Hydroxyflavanone: A novel strategy for targeting breast cancer. Oncotarget 2017;8:75025-37. [Crossref] [PubMed]
- Dan C, Zhang H, Zeng W, et al. HNF1B expression regulates ECI2 gene expression, potentially serving a role in prostate cancer progression. Oncol Lett 2019;17:1094-100. [PubMed]
- Wu R, Tang S, Wang M, et al. MicroRNA-497 Induces Apoptosis and Suppresses Proliferation via the Bcl-2/Bax-Caspase9-Caspase3 Pathway and Cyclin D2 Protein in HUVECs. PLoS One 2016;11:e0167052. [Crossref] [PubMed]
- Tan WL, Tan DS. Targeting the metastatic niche through anti-angiogenic approaches in epidermal growth factor receptor mutant non-small cell lung cancer. Transl Lung Cancer Res 2018;7:S13-8. [Crossref] [PubMed]
- Oudin MJ, Jonas O, Kosciuk T, et al. Tumor Cell-Driven Extracellular Matrix Remodeling Drives Haptotaxis during Metastatic Progression. Cancer Discov 2016;6:516-31. [Crossref] [PubMed]
- Wang J, He C, Gao P, et al. HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene 2020;39:1335-46. [Crossref] [PubMed]
- Malanga D, Belmonte S, Colelli F, et al. AKT1E17K Is Oncogenic in Mouse Lung and Cooperates With Chemical Carcinogens in Inducing Lung Cancer. PLoS One 2016;11:e0147334. [Crossref] [PubMed]
- De Marco C, Laudanna C, Rinaldo N, et al. Specific gene expression signatures induced by the multiple oncogenic alterations that occur within the PTEN/PI3K/AKT pathway in lung cancer. PLoS One 2017;12:e0178865. [Crossref] [PubMed]
- Pompura SL, Dominguez-Villar M. The PI3K/AKT signaling pathway in regulatory T-cell development, stability, and function. J Leukoc Biol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Singh K, Pruski MA, Polireddy K, et al. Mst1/2 kinases restrain transformation in a novel transgenic model of Ras driven non-small cell lung cancer. Oncogene 2020;39:1152-64. [Crossref] [PubMed]
- Dang Z, Shangguan J, Zhang C, et al. Loss of protocadherin-17 (PCDH-17) promotes metastasis and invasion through hyperactivation of EGFR/MEK/ERK signaling pathway in hepatocellular carcinoma. Tumour Biol 2016;37:2527-35. [Crossref] [PubMed]
- Vega-Benedetti AF, Loi E, Moi L, et al. Clustered protocadherins methylation alterations in cancer. Clin Epigenetics 2019;11:100. [Crossref] [PubMed]


