Establishment and application of a method of next generation sequencing of 285 genes in lung cancer based on Ion-Proton platform
Introduction
Clinical oncology has already entered the era of personalized medicine or precision medicine, which needs right drug for the right patient at right time especially in targetable cancers harboring driver genes like EGFR, ALK and ROS1, etc. (1,2). Since the proposal of “precision medicine” plan in 2015 by US president Obama, next generation sequencing (NGS) develops quickly and has been applied extensively as a novel genetic screening and diagnostic technique for clinical detection. Its genetic characteristics for disease detection have become an important part of modern precision medicine (3-5). NGS has become an effective and acceptable method for clinical gene detection (6,7). Although it is still in the early stage of clinical application for the diagnosis and treatment of tumors (8-10), its continuous innovation had accelerated the people’s cognition on genetics markers and the molecular mechanisms of diseases. Genome alterations play a significant role in disease development and progression a management by predictive and biomarkers, identifying clinical decision making of targeted therapy. The initial sample of NGS can be genomic DNA (gDNA) or circulating free DNA (cfDNA). Liquid biopsy refers to targetable alterations or other biomarkers using circulating material like cfDNA, CTC and CSF, etc. (11,12). Given that many clinically relevant biomarkers continue to be identified, and targeted drugs are developed for personalized treatment, multigene mutation screening will be a requirement in routine clinical practice (13). Genetic mutations can be detected by Sanger sequencing, gene chips, super ARMS, digital polymerase chain reaction (PCR), FISH and immunohistochemistry (IHC). However, these method presents several limitations. For instance, all the methods can only detect genetic mutations that have been found of very few numbers of genes. A large number of genetic variants that may be related to the disease can not be detected. Therefore, a multigene detection method that is suitable for Chinese lung cancer patients should be established.
Briefly, the motivations of developing this new method are as follows: (I) NGS can simultaneously interrogate multiple gene mutation loci, which are screened for in-routine clinical practice. (II) Tissue specimens and nucleic acids are not enough to used for an analysis of multiple genes one after another. However, NGS method can reduces the amount of the clinical sample used. (III) Some novel molecular targets of Chinese lung cancer patients can be enrolled in the new method. (IV) The NGS method can detect a variety of genetic variants, such as single nucleotide polymorphism (SNP), InDel, Fusion and copy number variation (CNV).
We developed this customized NGS to detected a total of 285 cancer related genes and applied to cell lines and clinical specimens for the validation of its performance.
We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2855).
Methods
Cancer cell lines, cancer tissues and patient selection
We used four NSCLC cell lines (i.e., H1650, H1975, A549 and H1299) that were selected from the cell bank of Guangdong Lung Cancer Institute (GLCI) for LoD (limit of detection) analysis of serial dilution of gDNA of these cells at 1:1, 1:5, 1:10 and 1:100. Twenty-four tumor tissues and 1 plasma was collected from the GLCI of Guangdong General Hospital between January 2015 and December 2016. All tissues samples were stored at –80 °C after being frozen in liquid nitrogen, were routinely assessed by pathologists to ensure that >20% tumor contend in each sample to be tested by NGS. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Guangdong Provincial People’s Hospital [No. GDREC2015360H(R2), GDREC2016316H(R1)]. All patients were well informed and signed the informed consent.
Reagents and instruments
The following materials were used in this study: QIAGEN QlAampDNA Mini Kit (Qiagen, Valencia, Germany); QIAGEN QlAampBlood Mini Kit (Qiagen, Valencia, Germany); Ion Xpress Plus Fragment Library Kit (Life Technologies, New York, USA); Agencourt AMPure XP Kit (Beckman, USA); H2O (Sigma-Aldrich, USA); Thermo NanoDrop 1000 (Thermo, MA, USA); SureSelect Target Enrichment System for Sequencing on Ion-Proton (Agilent Technologies, USA) ; SureSelect TE Reagent Kit, PTN (Agilent Technologies, USA); Herculase II Fusion DNA Polymerase kit (Agilent Technologies, USA); Ion OneTouch 2 System (Life Technologies, New York, USA); Qubit 2.0 (Life Technologies, New York, USA); Ion PI Template OT2 200 Kit v2 (Life Technologies, New York, USA); Ion PI Sequencing 200 Kit v2 (Life Technologies, New York, USA); ABI 3730xl Sequencing Machine (Life Technologies, New York, USA); and PCR machine (Life Technologies, New York, USA); QIAxcel Bioanalyzer (Qiagen, Valencia, Germany); ABI Ion-Proton Sequencing Machine (Life Technologies, New York, USA).
Design of a panel of targeted 285 genes
Selection of related driver genes in lung cancer
Search for the latest literatures by retrieving the PubMed database, comparing with the information of gene mutation in databases like DrugBank, COSMIC, dbSNP, OMIM, ClinVar, 5,000 exomes and 1,000 genomes. A total of 285 genes related to lung cancer pathogenesis, chemoradiotherapy, targeted drug resistance and metastasis were determined and included as a set for probe library design and target sequencing.
Probe Library design for hybrid selection
The targeted hybridization probes library of 285 genes were designed using SureDesign software web site based on genome hg19/GRCh37 (https://earray.chem.agilent.com/suredesign/index.htm). Runned the software, introduced the 285 gene list and adjusted the parameters such as databases, target region, region extension and allow synonyms. The total length of target capture sequence was 0.967 Mb. All the 4,700 exon loci of the 285 genes were included in the probe design kit. Each probe kit contains 16 tests as requested by ourselves. The designed gene hybridization probe was synthesized by the Agilent company.
gDNA purification and quantification
The gDNA was isolated from each tissue sample (tumor content more than 20%) with the QIAGEN QIAampDNA Mini Kit and the circulating DNA was isolated from 4 mL plasma with the QIAGEN QIAampBlood Mini Kit. DNA quantitative analysis was finished by using the Qubit analyzer.
NGS library construction
NGS libraries were prepared from plasma DNA and tumor and cell line gDNA. For patient plasma DNA, 112 ng DNA were used for library construction without additional fragmentation. For tumor, and cell line gDNA, 200–1,000 ng DNA was sheared prior to library construction with Ion Xpress™ Plus Fragment Library Kit for 150 bp fragments. The NGS libraries were constructed using the Ion Xpress Plus Fragment Library Kit (Life Technologies, New York, USA). The cleanup steps was using the Agencourt AMPure XP Kit (Beckman, USA). Ligation was performed for 15 min at 25 °C and 5 min at 72 °C using Ion Xpress Barcode Adapters. Single-step size selection was performed by adding 140 µL (1.4X) of Agencourt AMPure XP beads to enrich for ligated DNA fragments. The ligated fragments were then amplified using Herculase II Fusion DNA Polymerase (Agilent Technologies, USA) and 9–11 PCR cycles, depending on input DNA mass. Library purity and concentration was assessed by spectrophotometer (Qubit 2.0) and qPCR (KAPA Biosystems), respectively. Fragment length was determined on a QIAxcel Bioanalyzer using the QIAxcel DNA screening Kit (Qiagen).
Hybrid capture selection of target regions and NGS
SureSelect Target Enrichment System for Sequencing on Ion-Proton was used according to the manufacturer's protocol. One library was included in a single capture hybridization. Following hybrid selection, the captured DNA fragments were amplified with 9 to 11 cycles of PCR using 5× Herculase II Rxn Buffer and 2 µL SureSelect PTN PCR primer mix and 1 µL Herculase II Fusion DNA Polymerase in 50 µL reactions. Multiplexed libraries were mixed by 5 libraries with 20 µL of 12 pM and were sequenced using Ion PI Sequencing 200 Kit v2 runs on the Ion-Proton.
Data analysis and interpretation
Upon sequence completion, the sequencing data were mapped to the hg19 reference genome with the Suite software (Life Technologies, version 5.0.2), and mutations were Called with the Variant Caller software (Life Technologies, version 5.0.2.1), and the VCF files was producted at the same time. The VCF files was uploaded to the Ion Reporter cloud analysis platform (Life Technologies, https://ionreporter.lifetechnologies.com), and gene mutation informations were annotated by the Ion Reporter with a series of filters. The variant filters include minimum quality, minimum coverage, maximum strand bias, maximum variant signal shift and so on. Integrative Genomics Viewer (IGV) software was applied by researchers to annotate the gene mutation information by comparing with the databases DrugBank, COSMIC, dbSNP, OMIM, ClinVar, 5,000 exomes and 1,000 genomes.
Results
Selection of 285 genes as a testing panel
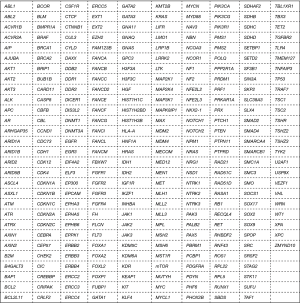
We check the cancer gene census through literatures and make a list of 285 genes including those common genes in lung cancer to be tested by NGS. We tested 4,700 regions in these 285 target genes, which were closely related to the pathogenesis, drug resistance, and metastasis of lung cancer and associated with relevant transduction pathways. The targeted 285 genes are listed in Figure 1.
NGS-related technical indicators
The proposed method was able to detect mutations of 285 genes in lung cancer cell lines and clinical lung cancer specimens. The reads, mapped reads, on target, mean depth and uniformity were 14.90±4.37 (×106), 98.68%±0.61%, 60.49%±10.72%, 714.42±264.13 and 90.51%±6.91%, respectively.
Establishment and validation of the customized NGS test in cell lines
The detection method used with the SureSelect target enrichment system for sequencing kit was established by analyzing four cell lines of lung cancer. The results from the newly established method were consistent with the previously reported mutations in cell lines. All the results were confirmed by Sanger sequencing, and the sequencing results are shown in Figure 2. The detailed mutation sites within genes and proteins are listed in Table 1. For the serial dilution of gDNA of these four cell lines, mutations were successfully detected as shown in Table S1.
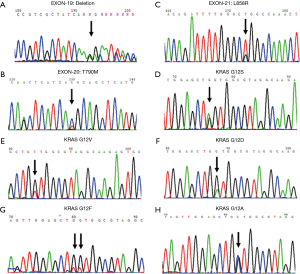
Table 1
| Cell line | Documented by ATCC | Mutations detected by established method |
|---|---|---|
| H1650 | EGFR_ Exon19 deletion | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) |
| H1975 | EGFR_L858R; EGFR_T790M | EGFR_L858R (c.2573T>G); EGFR_T790M (c.2369C>T) |
| A549 | KRAS mutation | KRAS_G12S (c.34G>A) |
| H1299 | EGFR/ALK/KRAS negative | No mutation |
Comparison of the customized NGS test and Sanger sequencing in lung cancer specimens
A total of 13 lung cancer tissue specimens were detected by the established method and Sanger sequencing. This method was validated through comparison by the Sanger sequencing. The results show that established method can detected all the mutations analyzed by the Sanger sequencing. ERBB2_E770delinsEAYVM, MET_S701N, PDGFRA_T674I, TP53_G245V, TP53_V274A, TP53_A276F, TP53_G334L, TP53_R337L and TP53_Y220C mutations were detected only through the established method. All the mutations results are shown in Table 2. All the Sanger sequencing results and established method results are shown in Figures 2,3, respectively.
Table 2
| Sample ID | Mutations detected by established method | MAF, % | Mutations detected by Sanger method | Mutations detected by LungCarta | Fusion proteins or mRNA tested by IHC/RT-PCR method |
|---|---|---|---|---|---|
| 1162 | EGFR_L858R (c.2573T>G) | 6.8 | EGFR_L858R | – | – |
| EGFR_T790M (c.2369C>T) | 11.6 | ||||
| 29001 | KRAS_G12V (c.35G>T) | 9.6 | EGFR_T790M | – | – |
| 29003 | KRAS_G12F (c.34_35delGGinsTT) | 41.2 | KRAS_G12V | – | – |
| TP53_G245V (c.734G>T) | 42.8 | ||||
| 29005 | EGFR_L858R (c.2573T>G) | 38.4 | KRAS_G12F | – | – |
| TP53_V274A (c.821T>C) | 54.9 | ||||
| 29006 | EGFR_L858R (c.2573T>G) | 15.5 | EGFR_L858R | – | – |
| TP53_A276F (c.826_827delGCinsTT) | – | ||||
| ERBB2_E770delinsEAYVM (c.2310_2311insGCATACGTGATG) | 16.2 | ||||
| 29014 | KRAS_G12A (c.35G>C) | 47.8 | EGFR_L858R | – | – |
| 29015 | EGFR_L858R (c.2573T>G) | 16.9 | KRAS_G12A | – | – |
| EGFR_T790M (c.2369C>T) | 19.1 | ||||
| 29017 | KRAS_G12D (c.35G>A) | 29.4 | EGFR_L858R | – | – |
| MET_S701N (c.2102G>A) | 9.6 | ||||
| PDGFRA_T674I (c.2021C>T) | 8.5 | ||||
| 29018 | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) | 15.2 | EGFR_T790M | – | – |
| EGFR_T790M (c.2369C>T) | 11.6 | ||||
| 29029 | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) | 18.2 | KRAS_G12D | – | – |
| TP53_G334L (c.1000_1001delGGinsTT) | 25.6 | ||||
| 29035 | EGFR_p.Ser752_Ile759del (c.2253_2276delATCTCCGAAAGCCAACAAGGAAAT) | 49.7 | EGFR_Exon19 del | – | – |
| 29036 | EGFR_L858R (c.2573T>G) | 3.3 | – | – | – |
| TP53_R337L (c.1010G>T) | 25.7 | ||||
| 29037 | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) | 41.8 | EGFR_T790M | – | – |
| EGFR_T790M (c.2369C>T) | 11.4 | ||||
| TP53_Y220C (c.659A>G) | 53.6 | ||||
| 22840 | EGFR_L858R (c.2573T>G) | 32.9 | – | EGFR_L858R; EGFR_ A289V PIK3CA_ p.E542K |
– |
| EGFR_ A289V (c.866C>T) | 38.6 | ||||
| PIK3CA_ p.E542K (c.1624G>A) | 49.5 | ||||
| K1736T | EGFR_L858R (c.2573T>G) | 24.9 | – | EGFR_L858R MET_N375S |
– |
| MET_N375S (c.1124A>G) | 18.6 | ||||
| NFE2L3_p.Ser511_Pro513del (c.1529_1537delCTTCTGAAC) | 12.3 | ||||
| K1744T | EGFR_p.Glu746_Ala750del (c.2236_2250del15) | 71.76 | – | EGFR_Exon19 del | – |
| K1745T | No mutation | – | – | No mutation | – |
| K1746T | No mutation | – | – | No mutation | – |
| 1215 | EML4-ALK | 11.04 | – | – | ALK fusion (IHC) |
| 33071 | EML4-ALK | 6.4 | – | – | ALK fusion (IHC) |
| 22968 | EML4-ALK | 5.7 | – | – | ALK fusion (IHC) |
| 17001 | KIF5B-RET | 7.7 | – | – | KIF5B-RET (RT-PCR) |
| 1146 | KIF5B-RET | 12.9 | – | – | KIF5B-RET (RT-PCR) |
MAF, minor allele frequency; IHC, immunohistochemistry; RT-PCR, real-time polymerase chain reaction.
Comparison of the customized NGS test with LungCarta method in lung cancer patients
Five lung cancer tissue specimens were detected by the established method used the SureSelect target enrichment system for sequencing kit and LungCarta method. The results show that established method can detected all the mutations analyzed by the LungCarta method. The results of the established method and the LungCarta method were shown in Table 2. However, a p.Ser511_Pro513del mutation in NFE2L3 gene of K1736T was detected only through the established method (Figure 4).
Comparison of established method and IHC/real-time PCR (RT-PCR) method in lung cancer specimens
For ALK and RET fusion testing, five lung cancer tissue specimens were detected by the established method and IHC/RT-PCR method. The congruence rate of detect result of specimens between the established method and IHC method was 100%. The results are shown in Table 2 (Figure 5).
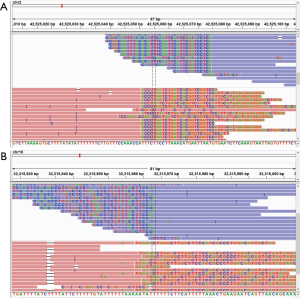
NGS method detected mutations in cancer tissues and plasma in a representative patient
Cancer tissue and blood from a special patient were detected by the established method. The detected results showed that the mutation information of the patient's tissue samples were identical to those in the blood sample. EGFR_T790M and Glu746_Ala750 del (c.2236_2250delGAATTAAGAGAAGCA) and TP53_Y220C (c.659A>G) mutations were detected in cancer tissues and blood. The results are shown in Table 3.
Table 3
| Patient | Mutations | |
|---|---|---|
| Cancer tissue (29037) | Plasma (29037-1) | |
| 29037 | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) |
| EGFR_T790M (c.2369C>T) | EGFR_T790M (c.2369C>T) | |
| TP53_ Y220C (c.659A>G) | TP53_ Y220C (c.659A>G) | |
Discussion
Single-gene detection methods for the molecular classification of lung cancer mainly include Quantitative PCR, Sanger sequencing, fluorescence in situ hybridization, amplification refractory mutation system, digital PCR and IHC. Multiplex genetic mutation-detection method include MassARRAY and gene chip. These methods can only detect the known gene mutations, and they are also expensive and time consuming. Many studies have shown tumors was complex, that tumors are not only associated with single driven gene mutations, but also associated with double or even multiple mutations (14). Considering the limitations of single-gene detection techniques and the complexity and unknowability of tumor driver genes, we need to establish a multiplex genetic mutation-detection method for lung cancer in clinical practice and translational medicine, Such as NGS. NGS has become an effective and acceptable method for clinical gene detection and has been an important molecular diagnostic tool for precision oncology (15,16).
According to the review of the related literature and data on lung cancer treatments, 285 genes were enrolled into the targeted panel. These genes not only include the eight genes which can using the related TKI of FDA-approved, such as EGFR, ALK, KRAS, ROS1, ERBB2, MET, RET and BRAF, but also include many tumor suppressors, such as TP53 gene. So, the established method can not only detect common L858R mutations in EGFR gene, but also detect recent reports of C1156Y, L1196M, F1174L, and G1269S mutations in ALK gene, which was recently reported as the resistance mutations following crizotinib treatment in lung cancer (17,18). The study found established method could detect the mutations verified by Sanger sequencing, also could detect the rare mutation, such as the p.Ser511_Pro513del mutation in NFE2L3 gene, that cannot be detected by the Sanger sequencing and MassARRAY platform. The results suggest that Compared to Sanger sequencing and MassARRAY, the established method has a high detection sensitivity and specificity. It is well known that the detection sensitivity of MassARRAY and Sanger were 1–5% (19) and 5–10% (20), respectively. So, the sensitivity of the established method should be less than 1%, that will be verified by the next phase of the experiment.
The proposed 285 genes-targeted NGS method demonstrates the following advantages: (I) the establishment NGS method can minimizes the weaknesses of traditional single-gene tests, such as time-consuming and tedious procedures and high cost. (II) Given the NGS library was built by a large number of fragmented DNA, theoretically, not only fresh tissue specimens but also formalin-fixed paraffin-embedded (FFPE) specimens, biopsy samples, pleural effusion and plasma could be used for effective detection, which needs further validation. (III) The establishment method can not only detect the known gene mutations, but also can detect the unknowability gene mutations.
Limitations of the study included relatively small sample size, absence of association analysis with clinical therapy. Thus, was provided about how the results of this analysis might affect clinical outcomes. Our study did not systematically measure the turn-around time required for NGS experiments and to annotate, interpret and report NGS results. Future studies will be necessary to address these limitations.
In summary, we have successfully established a customized novel NGS detection method based on Ion-Proton platform, which are able to detect those actionable driver gene mutations, and may significantly reduce the costs of biomarker tests and thus demonstrates a promising potential for personalized treatment.
Table S1
| Cell line | Serial dilution | Specific mutations | MAF, % |
|---|---|---|---|
| H1650 | 1:1 | EGFR_p.Glu746_Ala750del (c.2236_2250delGAATTAAGAGAAGCA) | 48.9 |
| 1:10 | 6.3 | ||
| 1:100 | 0.8 | ||
| H1975 | 1:1 | EGFR_L858R (c.2573T>G)/EGFR_T790M (c.2369C>T) | 56.8/49.3 |
| 1:10 | 6.8/5.9 | ||
| 1:100 | 1.1/0.9 | ||
| A549 | 1:1 | KRAS_G12S (c.34G>A) | 63.5 |
| 1:10 | 8.2 | ||
| 1:100 | 1.2 | ||
| H1299 | 1:1 | No mutation | – |
| 1:10 | – | ||
| 1:100 | – |
gDNA, genomic DNA; MAF, minor allele frequency.
Acknowledgments
Funding: This work was supported by following grants:
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at available at http://dx.doi.org/10.21037/tcr-19-2855
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2855
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2855). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Guangdong Provincial People’s Hospital [No. GDREC2015360H(R2), GDREC2016316H(R1)] and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol 2018;15:183-92. [Crossref] [PubMed]
- Schilsky RL. Implementing personalized cancer care. Nat Rev Clin Oncol 2014;11:432-8. [Crossref] [PubMed]
- Yang Y, Muany DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502-11. [Crossref] [PubMed]
- Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014;312:1880-7. [Crossref] [PubMed]
- Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA 2014;311:1035-45. [Crossref] [PubMed]
- Hayes DN, Kim WY. The next steps in next-gen sequencing of cancer genomes. J Clin Invest 2015;125:462-8. [Crossref] [PubMed]
- Garraway LA. Genomics-driven oncology: Framework for an emerging paradigm. J Clin Oncol 2013;31:1806-14. [Crossref] [PubMed]
- Good BM, Ainscough BJ, McMichael JF, et al. Organizing knowledge to enable personalization of medicine in cancer. Genome Biol 2014;15:438. [Crossref] [PubMed]
- Griffith M, Miller CA, Griffith OL, et al. Optimizing cancer genome sequencing and analysis. Cell Syst 2015;1:210-23. [Crossref] [PubMed]
- Hyman DM, Solit DB, Arcila ME, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today 2015;20:1422-8. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Tian HX, Zhang XC, Wang Z, et al. Establishment and application of a multiplex genetic mutation-detection method of lung cancer based on MassARRAY platform. Cancer Biol Med 2016;13:68-76. [Crossref] [PubMed]
- Yang JJ, Zhang XC, Su J, et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res 2014;20:1383-92. [Crossref] [PubMed]
- Motro Y, Moran-Gilad J. Next-generation sequencing applications in clinical bacteriology. Biomol Detect Quantif 2017;14:1-6. [Crossref] [PubMed]
- Serratì S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther 2016;9:7355-65. [Crossref] [PubMed]
- Heuckmann JM, Hölzel M, Sos ML, et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res 2011;17:7394-401. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Kriegsmann M, Arens N, Endris V, et al. Detection of KRAS, NRAS and BRAF by mass spectrometry-a sensitive, reliable, fast and cost-effective technique. Diagn Pathol 2015;10:132. [Crossref] [PubMed]
- Jancik S, Drabek J, Berkovcova J, et al. A comparison of direct sequencing, pyrosequencing, high resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res 2012;31:79. [Crossref] [PubMed]