
Comparison of whole exome sequencing in circulating tumor cells of primitive and metastatic nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common cancers in humans and frequently has a history of Epstein-Barr virus (EBV) infection associated with it and is prone to metastasize to distant lymph nodes and organs (1). The occurrence of NPC is a complex process that involves a combination of viral infection, environmental factors, and genetic aberrations (2,3). It is also related to diet habits, including the consumption of salted fish (4). In addition, recent studies have revealed that alterations of NF-κB signal pathway genes are also strongly associated with the pathology of NPC (5-7). The treatment of NPC patients currently consists of combination of radiotherapy and chemotherapy, including the use of cisplatin, 5-fluorouracil (5-FU), paclitaxel, and gemcitabine (7-9). However, the prognosis of most NPC patients is still poor. Therefore, applying genetic screening to detect specific biomarkers for treatment may be a better option, especially for patients with metastasis. To date, some reports have shown that soluble programmed death-ligand1 (sPD-L1) (10-12), microRNAs BART7-3p, BART13-3p (13,14), amyloid beta 4 (A4) (15), and soluble MHC class I chain-related molecule A (MICA) (16) are good candidates as NPC biomarkers. However, the sensitivity and specificity of these biomarkers are controversial. Therefore, many studies have tried to explore new specific methods for NPC diagnosis and outcome predictions, including the clinical significance of circulating tumor cells (CTCs) in NPC patients (17,18). To date, less NPC genomic data are available, which obstructs the understanding of NPC biology, disease progression, and selective treatment.
Next generation sequencing (NGS) is a very sensitive and reliable technique for disclosing complex genetic aberrations in cancer patients (19). Chow et al. (8) found that there is a high percentage of gene mutations in fresh NPC samples, including alterations in the EGFR-PI3K-Akt-mTOR, Notch, NF-κB, and DNA damage and repair (DDR) signaling pathways. Lin et al. (20) reported that frequent genetic lesions in NPC patients were closely associated with chromatin modification, autophagy, and the ERRBB-PI3K signaling pathway. These reports mainly utilized primitive tumor cells as sequencing sources, which may have limited their outcomes. Recent studies have indicated that whole exome sequencing (WES) from patient’s CTC samples reflect more accurately the genomic characterization of real tumors (21-23).
Here, we compared the gene mutations profiles of tumor cells and CTCs in primitive and metastatic NPC patients. CTCs originate from primitive tumors, where they are shed into the vasculature and/or lymphatics traveling in the blood circulation (24) to distant organs causing tumor metastasis. Therefore, the detection of CTC’s genomic DNA alterations is very helpful to determine patient prognosis and the appropriate treatment.
Methods
Subjects
We collected and sequenced a total of 12 samples from four patients from June, 2017 to June, 2018 at our hospital. Their identification numbers were K06275, 47 year-old male; K06269, 54 year-old male; K05734, 62 year-old male; K06262, 54 year-old male. K06275 and K06269 were patients with non-metastatic NPC. K06262 and K05734 were patients with metastasis. Samples were taken from peripheral white blood cells, primitive tumors using surgery or endoscopy, and CTCs. This study protocol was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by ethical committee of the People’s Hospital of Guangxi Zhuang Autonomous Region. Approved protocol number was 2017-23. All patients gave informed consent.
Samples DNA Extraction and CTCs isolation
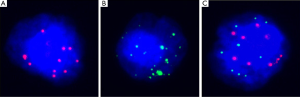
DNA from the primitive lesions in paraffin embedded tumors was extracted using the Maxwell16 FFPE plus LEV DNA purification Kit (Promega, Madison, USA). For CTC isolation, we followed the protocols described as the previous paper (25). Briefly, 5 mL peripheral blood from the patient’s vein was first lysed using red blood lysis buffer and then filtered through an 8 µM filter membrane. CTCs remained on the filter membrane, whereas white blood cells passed through the filter, after which, their DNA was extracted using the Maxwell 16 cell LEV DNA Purification kit (Promega, Madison, USA). The membrane with CTCs was hybridized using CanPatrolTM CTC RNAISH (SurExam Bio-Tech, Guangzhou, China) and identified. Then, confirmed CTCs were collected using a Palm Microbeam laser microdissection system and further enriched and amplified with the GenomePlex Single Cell. CTCs were classified as epithelial, mesenchymal, and mixed types according to their morphology and surface markers. Their graphs were shown as Figure 1. Whole Genome Amplification Kit (Sigma, USA). Finally, DNA fragments were obtained from amply and purification and were used for all subsequent experiments.
DNA library generation and sequencing
The above mentioned DNA was processed for amplification of targeting regions, primers digestion, adapter connection, fragment screening, and library enrichment with the Ion AmpliSeqTM Exome RDY 4×2 kit (Life Technologies, USA). Then, a library was constructed, preparing sequencing template with the use of Ion ChefTM (Life Technologies, USA). The sequencing templates were then transferred into PI sequencing chips and connected to an Ion Proton sequencing instrument (Life Technologies, USA) for high-throughput sequencing (HTS).
Experiment flowchart
The exome is the sum of all exons in one specimen. This region contains essential genetic information for protein translation (26). Exome sequencing utilized chips and probes hybridizing genomic deoxyribonucleic acid (DNA) sequencing of riched exons. Then, high throughout sequencing was-used to detect all samples.
Bioinformation analysis strategy
Raw reads were obtained from WES. Then, low quality (<10) sequences of less than 50 base pair were ruled out. After decontamination unique pairs were aligned with unique mapped reads in the whole genomic database. Subsequent genetic information analysis included targeting region sequencing depth, covering rate analysis, single-nucleotide polymorphism (SNP), and Indel detection, using the bioinformatics analysis tool Annovar. After screening for mutated genes with altered amino acid, we analyzed the linkage of the molecular mechanism, signal pathways and cellular functions in these altered genes and proteins using gene ontology (GO) analysis.
Results
Somatic mutational landscape in 4 NPC patients
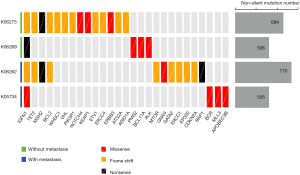
To compare the profile of gene mutations in primitive and metastatic NPC patients, WES of white blood cells, tumor cells and CTCs from patients was performed and analyzed. All 4 patients were male. The age was between 47 to 62 years old. After ion torrent analysis of raw data of sequencing, they were screened and filtered. Then, we carried out Annovar analysis. Non-exonic region mutation sites, adapter, non-monoclonal fragments (multiple barcode), and synonymous mutations (SNV) were first ruled out. We chose non-synonymous mutations (non-SNV) as our data for analysis. The one of following three criteria was selected as our potential candidate genes: (I) significantly mutated genes previously reported in a large scale sequencing of NPC patients (20); (II) relevant genes in NPC as well as other cancers listed in the COSMIC database (http://cancer.sanger.ac.uk/cosmic/) (27); (III) genes that act in-pathways related to cancer according to the KEGG database (https://www.genome.jp/kegg). According to these criteria, we obtained the mutational landscape of 4 primitive tumor lesions (Figure 2). We found no significant difference between the non-SNV of the primitive lesions in the metastatic patients (K06262 and K05734) and the non-metastatic patients (K06269 and K06275). However, we found that there was MSH2 gene mutation in two non-SNV patients (K06275 and K06262), which suggested that the MSH2 gene mutation give rise to defects of the DDR function. This process in turn may have promoted the accumulation of mutations involved in tumorigenesis. In addition, we also found BAP1 gene alteration in a metastatic patient (K06262), which is relevant to invasion and tumor metastasis (28). This result indicated that this gene mutation enhanced the invasive ability of cancer cells, and resulting in metastasis.
Mutational signature of the primary lesions and CTC samples
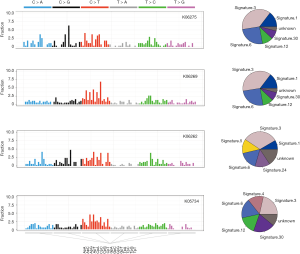
All single-nucleotide variations (SNVs) were classified into six categories according to the mutation directions (C > A, C > G, C > T, T > A, T > C, and T > G) (6). On the addition of mutated nucleotides at the 5’-terminal and 3’-terminal, there was total of 96 mutational contexts. Different mutational contexts have varying proportions in each tumor. This different ratio of 96 mutational contexts is defined as the mutational signature (29). Each signature has its corresponding generation mechanism. Therefore, we speculated that the mutational signature of each tumor contributes to its tumorigenesis. We analyzed a fraction of the 96 mutational contexts in four primitive NPC patients using the multiple linear regression models (30), and compared their mutational contexts with 30 mutational signatures in the COSMIC database (Figure 3). The left panel in Figure 3 shows a fraction of six mutational contexts and the right panel shows the-proportion of each signature. We found that all the four patients had a dominant 3 type signature, which results from double strand DNA break and causes dysfunction of homologous recombination. This also indicated that the genome is unstable in the process of carcinogenesis. Interestingly, we also found the unique signatures 5 and 4 in two metastatic patients, K06262 and K05734, respectively. The resulting mechanism of signature 5 is unclear; however, it has been identified in many kinds of cancers. In contrast, signature4 frequently occurs as a C > A transition in the transcribed strand that results from smoking. These results indicated that distinct life environments and styles caused different carcinogenesis mechanisms.
To evaluate specific gene mutations in CTCs, we also performed differential mutation analysis. Gene mutations from leukocytes were used as a negative control. We found almost identical mutations between primitive tumor and white blood cells. In contrast, there were tremendous differences between CTCs and white blood cells (Table 1). Specially, we found that CFAP74, MOB3C, PDE4DIP, IGFN1, CYFIP2, NOP16, SLC22A1, ZNF117 and SSPO mutations were involved in both primary tumor and CTC samples. CCDC144NL only occurred in primitive tumors. Interestingly, OR2T12, CPN2, MLXIPL, BAIAP3, IGSF3, SIN3B, and ZNF880 mutations were found in the metastatic group (supplementary material at http://fp.amegroups.cn/cms/123ade0661df7777fed15ae4e1cea7fa/TCR-19-2899-1.xlsx).
Table 1
| Gene | K06269 PT | K06275 PT | K06262 PT | K05734 PT | K06269 CTC | K06275 CTC | K06262 CTC | K05734 CTC |
|---|---|---|---|---|---|---|---|---|
| SAMD11 | cT257A | |||||||
| NOC2L | cT1532C, cG1531A, cA1529G, T672G | C2018_2020del, C1178_1179GA, cA92G | cA1024T, cG451A, c109_110del, cG67A | |||||
| KLHL17 | cA13G, G52A, cG1396A, cC1654A, cG1737C, cG1812A | cT1261C | cG1730C | cG1703A | ||||
| PLEKHN1 | cC1186T | cG568A, cA868G, cA1241G | ||||||
| PERM1 | cA2299G, cT2204C, cA1652C, cC1649T, cG1640C, cG1626T, cT1105C, cA1096G, cT1085C, CT58A | cG1357A, cC713T, cC707T, cC457T | ||||||
| ISG15 | cA188G | cG40A | ||||||
| AGRN | cG2404A, cC2423T, cC4688T, cA4696G, cA4697C, cG4924A, cG5883C | cG5536T, cG5626T | cT1732C, cA2066T, cG3421T, cG3433A, cG4324A, cG4456A | cT281C, cG1853A, cG2486A, cG2563A, cA2573G, cT2761C, cG4501A | ||||
| TTLL10 | cC969A | c731delC, cG740A | ||||||
| TNFRSF18 | cC562T | cC445T | ||||||
| TNFRSF4 | cT446C | cG269A | ||||||
| SDF4 | cG1015A, cG973A, cG763C, cG704A, cG657C, cG583A, cG280T, cG263A, cC259A, cC169A, cT143C | cC1022T, cC650T | ||||||
| UBE2J2 | cT247C | cT239G | cT527A | cT122C | ||||
| SCNN1D | cG163C | cG587A, cA1978G | ||||||
| ACAP3 | cG2204T, cG1789A, cG1733T, cG1637A, cT1616A, cG1475T, cA1474G, cG1468C, cG1432A | cC982T |
GO pathway enrichment analysis
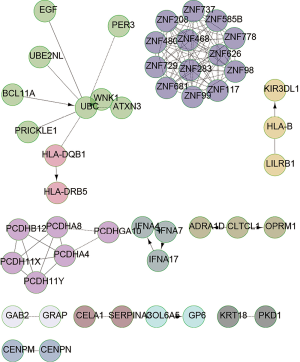
We further analyzed the pathway enrichment of gene mutations using GO analysis. The results are shown in the Figures 4 and 5 for the NPC patients. As an example, the graph shows that the UBC complex, ZNF family numbers, PCDH proteins, and IFNA were key mutated genes in patient K06269 patient (Figure 4). In contrast, PCDHGA10, SRA1, ZNF family numbers, CNOT1 and WNK1 gene mutations were involved in alterations of signaling pathways in patient K05734 (Figure 5). Similarly, HSPG2, NOTCH 4, PXN, PIK3R1 and GAB1 gene mutations occurred in patient K06275. CD40, HSPG2, EP300, LIMK1 and GNAS gene alteration were relevant to different signaling pathways in patient K06262 (data not shown). These data revealed that individual tumors had their own altered signaling pathways.
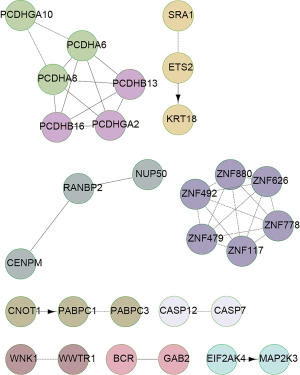
Profiles of cancer associated gene mutations
Carcinogenesis is a multi-step procession many gene mutations accumulate at various stages. Up to 400 genes including oncogenes, tumor suppressor, cell cycle related, apoptotic, RNA transcription and translation, DDR genes are involved in this transformation. The present study confirmed these findings in the metastatic patient K 06262 (Table 2). These critical gene mutations definitively contributed to the occurrence of NPC. We found PMS2, BCL11A, PDE4DIP, and ALK4 gene mutations in the K06269 primitive lesion, which are related to DDR, metabolism, cell proliferation, and protein degradation. In contrast, ERCC3, EGFR, BRAF, and 155 gene mutations occurred in K06269 CTC samples. Similarly, MSH2, ERCC4, VHL, and 21 more gene mutations occurred in the K06275 primary lesion; PARP1, ERCC5, APC, and 264 more gene mutations occurred in K06275 CTC samples. Interestingly, MSH2, ERCC1, BAP1, and 26 more gene mutations, and MSH2, ERCC3, EGFR, and 240 more gene mutations occurred in K06262 primitive lesion and CTC samples, respectively. PDE4DI, and BCR2 mutations occurred in the patient K05734 primitive lesion. MSH2, ERCC3, HIF1A, and 237 more gene mutations occurred in the patient K05734 CTC sample (Table 3). We found that K06275 always had more mutations than the other three subjects in either primitive lesions or CTC samples. In contrast, subject K06262 from the metastatic group had more tumor associated gene mutations. Only K06262 had invasion relevant gene mutations in primitive lesions. However, all four CTC samples had more invasion and metastasis relevant gene mutations than primitive lesions.
Table 2
| Gene | AAChange | ExonicFunc.refGene | Classification |
|---|---|---|---|
| MSH2 | c.196_197insG | Stopgain | DNA damage and repair |
| ERCC1 | c.211delG | Frameshift deletion | DNA damage and repair |
| GATA2 | c.441dupG | Frameshift insertion | RNA transcription and translation |
| TET2 | c.4582delC | Frameshift deletion | RNA transcription and translation |
| PML | c.1767delG | Frameshift deletion | RNA transcription and translation |
| JAK3 | c.G2774T | SNV | RNA transcription and translation |
| CRTC1 | c.1706delC | Frameshift deletion | RNA transcription and translation |
| CEBPA | c.206delC | Frameshift deletion | RNA transcription and translation |
| CEBPA | c.218_219del | Frameshift deletion | RNA transcription and translation |
| CEBPA | c.208_209del | Frameshift deletion | RNA transcription and translation |
| CEBPA | c.212delG | Frameshift deletion | RNA transcription and translation |
| EP300 | c.4402dupA | Frameshift insertion | RNA transcription and translation |
| PDE4DIP | c.T5207A | SNV | Metabolism |
| PDE4DIP | c.C2353T | Stopgain | Metabolism |
| PDE4DIP | c.1218delA | Frameshift deletion | Metabolism |
| PDE4DIP | c.G180A | Stopgain | Metabolism |
| RET | c.960_961del | Frameshift deletion | Metabolism |
| BCL9 | c.1148_1149insG | Frameshift insertion | Development and differentiation |
| ACVR2A | c.29_30insA | Frameshift insertion | Development and differentiation |
| ACVR2A | c.G31T | Stopgain | Development and differentiation |
| TNK2 | c.21delC | Frameshift deletion | Development and differentiation |
| HNF1A | c.A1720G | SNV | Development and differentiation |
| CIC | c.407_408insC | Frameshift insertion | Development and differentiation |
| IKBKB | c.151delC | Frameshift deletion | Immunology |
| CDKN2A | c.342delC | Frameshift deletion | Apoptosis |
| BCL2L2 | c.265delG | Frameshift deletion | Apoptosis |
| EZH2 | c.1682delA | Frameshift deletion | Proliferation |
| BCL2 | c.119_120del | Frameshift deletion | Proliferation |
| TCF3 | c.1474delG | Frameshift deletion | Proliferation |
| MARK4 | c.1310delG | Frameshift deletion | Proliferation |
| MTOR | c.3041_3042insT | Frameshift insertion | Signal transduction |
| GNAS | c.C1522A | SNV | Signal transduction |
| BAP1 | c.G88T | Stopgain | Tumor metastasis |
Table 3
| Name | Classification | K06269 PT | K06275 PT | K06262 PT | K05734 PT | K06269 CTC | K06275 CTC | K06262 CTC | K05734 CTC |
|---|---|---|---|---|---|---|---|---|---|
| BRIP1 | DNA damage | cT1904C | cC2443T, cA394C | ||||||
| DEE | DNA damage | C795delA, C737del | |||||||
| ERCC1 | DNA damage | C211delG | cG^10A | ||||||
| ERCC2 | DNA damage | cT1987C, cA1936C | cA523G | ||||||
| ERCC3 | DNA damage | cG421A, cA206G | cG163A | cC1611A | |||||
| ERCC4 | DNA damage | C1107delA | cG610A | ||||||
| ERCC5 | DNA damage | cA806G, cG2060A | cG1109A, c1232delC | cA1333C, cC2698T | |||||
| FANCA | DNA damage | cA2069G, cT1171C | cT3322C | ||||||
| FANCD2 | DNA damage | cG659A, cG898C | cT328C, cC1382A | cA887T, cA2420G | cC221T, cT2245A | ||||
| FANCG | DNA damage | cG248A, cT206C | cT866C, cG259A | cC1216T | |||||
| MLH1 | DNA damage | cT259A, cC671T | cA1138G | cG395A, cT578C | |||||
| MRE11A | DNA damage | cT851C, cA517G | cT466C | ||||||
| MSH2 | DNA damage | C196-197invG | C196-197invG | cA37G, cT50C | cC1799T, cG1862A | cT2269A | cG847A, cT895C | ||
| MSH6 | DNA damage | cG421C, cC587T | cA2367C, cT2959C | cG2689A, cA2822G | |||||
| MUTYH | DNA damage | cA1583G, cT863C | cA155G | ||||||
| NBN | DNA damage | cG1609G | cT2245C, c1181del | ||||||
| NSD1 | DNA damage | cC73T, cA856C | cT3365C, cT4880C | cA527GcA1961C, | cT115C, cG634A | ||||
| PALB2 | DNA damage | cA2242T, cG829A | cT3215G, cT2593C | ||||||
| PARP1 | DNA damage | cA2108G, c1791delT | cA2173G, cT1982C | cG1480A, cG447C | cC2488A | ||||
| PMS1 | DNA damage | cC3017T, cG775C | cA133G, cA976G | C518delA | cG1390A, cT1652C | ||||
| PMS2 | DNA damage | cC1454A | cT866C | ||||||
| RAD50 | DNA damage | cG1131T, cA1192G | cC2354T, cA2930G | cA2567G, cT3664C | cA140G, c163delG | ||||
| RECQL4 | DNA damage | cG3552A, cT2282C | cC1415T | cT3083G, cC2309T |
CTCs, circulating tumor cells; NPC, nasopharyngeal carcinoma.
Comparison of gene mutations in CTC samples
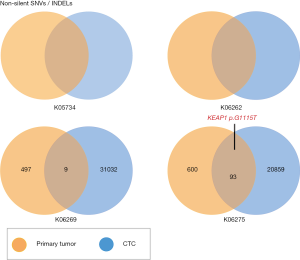
CTCs are considered one of the causes for cancer metastasis. Primitive tumor cells enter the peripheral blood and seed tumor cells into long distant organs, where they proliferate and amplify to form metastatic sites (31). We isolated CTCs from four patients using CTC isolation kits and performed WES to compare them with primitive tumors. The results are shown in Figure 6. We detected large amounts of somatic mutations. Although there were some identical mutations between primitive tumors and CTCs, we found that somatic mutations in CTCs were considerably higher than in the primitive tumor. These results indicated that CTC WES is very helpful tumor prediction and for diagnosis.
Discussion
Carcinogenesis is involved in the alteration of multiple molecules and signaling pathway networks (32,33). Activation of oncogenes or inactivation of suppressor genes can give rise to the occurrence of cancers (34,35). Here, we utilized WES to trace gene alterations in primitive tumors and CTCs of two non-metastatic patients and two metastatic patients. The present data revealed that the somatic mutation rate in CTCs is significantly higher than in either primitive tumor or metastatic samples. Interestingly, the patients with high non-synonymous mutations had MSH2 gene mutation. In addition, we also found that BAP1 gene alteration, which is relevant to invasion and metastasis of cancer, occurred in a metastatic patient.
MSH2 gene encodes a DNA mismatch repair (MMR) protein, which is involved in many kinds of DNA repair (36). MSH2 alteration is frequently associated with hereditary nonpolyposis colorectal cancer (HNPCC) because of DNA microsatellite instability (37). Interestingly, immunotherapy of colorectal cancer patients with MSH2 mutation has shown great benefits (38). Our qPCR results revealed that there were significant high PD-L1 and CTLA4 levels in metastatic patients compared with non-metastatic patients. Previous studies have shown extensive expression of PD-L1 had extensive expressions in tumor cells or immune cells of NPC patients (39,40). The other data also showed that there were high PD-L1 level in NPC patients with MMR status (41). All together, these finding indicate that the use of immune checkpoint inhibitors in metastatic NPC patients may result in great outcomes. MSH2 dimerizes with MSH6 to form the MutSα mismatch repair complex, which repairs DNA longer insertion/deletion loops (42). MSH2 alteration is also involved in acute lymphoblastoid leukemia (ALL) patients (43). Here, we found an association between MSH2 mutations was associated in primitive and metastatic NPC patients with high somatic mutation ratio. This implied a key role of DDR gene dysfunction in the occurrence of NPC.
BAP1 gene encodes a deubiquitinating enzyme that acts as a nuclear-localizing protein. Mutations in this gene have been identified in some breast and lung cancers (27). BAP1 mutation is closely relevant to metastasis (28). In the present result showed that BAP1 alteration only happened in metastatic patient, confirming its role in metastasis. Two metastatic patients had the unique mutational signatures 4 and 5. This indicated that each patient is unique with his own environment and life style.
To date, previous reports have shown that sPD-L1 (10-12), microRNAs BART7-3p, BART13-3p (13,14), A4 (15), and MICA (16) as candidate biomarkers for NPC prognosis. The clinical significance of these markers for NPC metastasis remains. Recent studies revealed that gene mutations in the NF-κB signaling pathway were involved in the occurrence of NPC (5,6,8,44). Our current data revealed that more gene mutation occurred in NPC patients, including those in DDR, RNA transcription and translation, metabolism, apoptosis, and immunology pathways. This result hinted that the occurrence of NPC is more complex than we previously thought. We also found that CFAP74, MOB3C, PDE4DIP, IGFN1, CYFIP2, NOP16, SLC22A1, ZNF117, and SSPO mutations were present in all primitive tumor and CTC samples. Interestingly, OR2T12, CPN2, MLXIPL, BAIAP, IGSF3, SIN3B and ZNF880 only occurred in metastatic NPC patients. This finding demonstrated that these genes may be used as new biomarkers to target therapy. We plan to test these mutated genes in large a sample size in the future.
We also found that many DDR relevant gene mutations including BRIP1, PMS1 and DEE only existed in metastatic CTC samples. Graham et al. (45) found that PMS1 forms a complex with MIH1gene to correct mispaired DNA. Zhao (46) reported that MLH1, MSH2, MSH6 and PMS1 are present in stage II and III colorectal cancer patients. Velázquez et al. (47) found that BRIP1 gene mutations occurred in inherited breast cancer patients. It was also reported that the association between MSH6 and BRIP1 variants are relevant to oxidative DNA damage in triple negative breast patients (48). Here, we first have found that PMS1 and BRIP1 gene mutations are relevant to NPC metastasis.
Conclusions
Our data revealed that WES of CTC samples in NPC patients is a very powerful tool for distinguishing primitive and metastatic tumor. We found that a few critical gene mutations, such as those in the MSH2, BAP1, PMS1, DEE and BRIP1 genes are present in metastatic CTC samples. These genes may be used as new biomarkers to target-treatment.
Acknowledgments
The authors would like to thank the support from the National Science Foundation of China and the Major Program of Science and Technology Development Project of Guangxi Province.
Funding: This study was supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2899
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2899). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study protocol was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by ethical committee of the People’s Hospital of Guangxi Zhuang Autonomous Region. Approved protocol number was 2017-23. All patients gave informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Banko AV, Lazarevic IB, Folic MM, et al. Characterization of the Variability of Epstein-Barr Virus Genes in Nasopharyngeal Biopsies: Potential Predictors for Carcinoma Progression. PLoS One 2016;11:e0153498. [Crossref] [PubMed]
- Zhang F, Zhang J. Clinical hereditary characteristics in nasopharyngeal carcinoma through Ye-Liang's family cluster. Chin Med J (Engl) 1999;112:185-7. [PubMed]
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol 2012;22:79-86. [Crossref] [PubMed]
- Yu MC, Ho JH, Lai SH, et al. Cantonese-style salted fish as a cause of nasopharyngeal carcinoma: report of a case-control study in Hong Kong. Cancer Res 1986;46:956-61. [PubMed]
- Li YY, Chung GT, Lui VW, et al. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat Commun 2017;8:14121. [Crossref] [PubMed]
- Zheng H, Dai W, Cheung AK, et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-kappaB pathway regulators in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2016;113:11283-8. [Crossref] [PubMed]
- Verhoeven RJ, Tong S, Zhang G, et al. NF-kappaB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J Virol 2016;90:6475-88. [Crossref] [PubMed]
- Chow YP, Tan LP, Chai SJ, et al. Exome Sequencing Identifies Potentially Druggable Mutations in Nasopharyngeal Carcinoma. Sci Rep 2017;7:42980. [Crossref] [PubMed]
- Bensouda Y, Kaikani W, Ahbeddou N, et al. Treatment for metastatic nasopharyngeal carcinoma. Eur Ann Otorhinolaryngol Head Neck Dis 2011;128:79-85. [Crossref] [PubMed]
- Lu T, Chen Y, Li J, et al. High Soluble Programmed Death-Ligand 1 Predicts Poor Prognosis in Patients with Nasopharyngeal Carcinoma. Onco Targets Ther 2020;13:1757-65. [Crossref] [PubMed]
- Wotman M, Herman SW, Costantino P, et al. The Prognostic Role of Programmed Death-Ligand 1 in Nasopharyngeal Carcinoma. Laryngoscope 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Minichsdorfer C, Oberndorfer F, Krall C, et al. PD-L1 Expression on Tumor Cells Is Associated With a Poor Outcome in a Cohort of Caucasian Nasopharyngeal Carcinoma Patients. Front Oncol 2019;9:1334. [Crossref] [PubMed]
- Lu T, Guo Q, Lin K, et al. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci 2020;111:1711-23. [Crossref] [PubMed]
- Wu L, Wang J, Zhu D, et al. Circulating Epstein-Barr virus microRNA profile reveals novel biomarker for nasopharyngeal carcinoma diagnosis. Cancer Biomark 2020;27:365-75. [Crossref] [PubMed]
- Li XY, Meng HL, Li KG, et al. Amyloid Beta (A4) Precursor Protein: A Potential Biomarker for Recurrent Nasopharyngeal Carcinoma. Cancer Manag Res 2019;11:10651-6. [Crossref] [PubMed]
- Ben Chaaben A, Ouni N, Douik H, et al. Soluble MICA and anti-MICA Antibodies as Biomarkers of Nasopharyngeal Carcinoma Disease. Immunol Invest 2020;49:498-509. [Crossref] [PubMed]
- Wen Z, Li Z, Yong P, et al. Detection and clinical significance of circulating tumor cells in patients with nasopharyngeal carcinoma. Oncol Lett 2019;18:2537-47. [PubMed]
- Xie XQ, Luo Y, Ma XL, et al. Clinical significance of circulating tumor cells and their expression of cyclooxygenase-2 in patients with nasopharyngeal carcinoma. Eur Rev Med Pharmacol Sci 2019;23:6951-61. [PubMed]
- Teng CF, Huang HY, Li TC, et al. A Next-Generation Sequencing-Based Platform for Quantitative Detection of Hepatitis B Virus Pre-S Mutants in Plasma of Hepatocellular Carcinoma Patients. Sci Rep 2018;8:14816. [Crossref] [PubMed]
- Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet 2014;46:866-71. [Crossref] [PubMed]
- Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479-84. [Crossref] [PubMed]
- Dai W, Zheng H, Cheung AK, et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2016;113:3317-22. [Crossref] [PubMed]
- Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526-31. [Crossref] [PubMed]
- Riquet M, Rivera C, Gibault L, et al. Lymphatic spread of lung cancer: anatomical lymph node chains unchained in zones. Rev Pneumol Clin 2014;70:16-25. [Crossref] [PubMed]
- Wu S, Liu S, Liu Z, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One 2015;10:e0123976. [Crossref] [PubMed]
- Gilbert W. Why genes in pieces? Nature 1978;271:501. [Crossref] [PubMed]
- Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998;16:1097-112. [Crossref] [PubMed]
- Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010;330:1410-3. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [Crossref] [PubMed]
- Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 2013;110:21083-8. [Crossref] [PubMed]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [Crossref] [PubMed]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer 2001;1:157-62. [Crossref] [PubMed]
- Vlahopoulos SA, Logotheti S, Mikas D, et al. The role of ATF-2 in oncogenesis. Bioessays 2008;30:314-27. [Crossref] [PubMed]
- Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science 2006;312:1650-3. [Crossref] [PubMed]
- Pitsikas P, Lee D, Rainbow AJ. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis 2007;22:235-43. [Crossref] [PubMed]
- Bonis PA, Trikalinos TA, Chung M, et al. Hereditary nonpolyposis colorectal cancer: diagnostic strategies and their implications. Evid Rep Technol Assess (Full Rep) 2007;1-180. [PubMed]
- Overman MJ, Ernstoff MS, Morse MA. Where We Stand With Immunotherapy in Colorectal Cancer: Deficient Mismatch Repair, Proficient Mismatch Repair, and Toxicity Management. Am Soc Clin Oncol Educ Book 2018;38:239-47. [Crossref] [PubMed]
- Larbcharoensub N, Mahaprom K, Jiarpinitnun C, et al. Characterization of PD-L1 and PD-1 Expression and CD8+ Tumor-infiltrating Lymphocyte in Epstein-Barr Virus-associated Nasopharyngeal Carcinoma. Am J Clin Oncol 2018;41:1204-10. [Crossref] [PubMed]
- Chan OS, Kowanetz M, Ng WT, et al. Characterization of PD-L1 expression and immune cell infiltration in nasopharyngeal cancer. Oral Oncol 2017;67:52-60. [Crossref] [PubMed]
- Zhao L, Liao X, Hong G, et al. Mismatch repair status and high expression of PD-L1 in nasopharyngeal carcinoma. Cancer Manag Res 2019;11:1631-40. [Crossref] [PubMed]
- Dowen JM, Putnam CD, Kolodner RD. Functional studies and homology modeling of Msh2-Msh3 predict that mispair recognition involves DNA bending and strand separation. Mol Cell Biol 2010;30:3321-8. [Crossref] [PubMed]
- Diouf B, Cheng Q, Krynetskaia NF, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med 2011;17:1298-303. [Crossref] [PubMed]
- Li Y, Qin Y, Yang C, et al. Cardamonin induces ROS-mediated G2/M phase arrest and apoptosis through inhibition of NF-kappaB pathway in nasopharyngeal carcinoma. Cell Death Dis 2017;8:e3024. [Crossref] [PubMed]
- Graham WJ, Putnam CD, Kolodner RD. The properties of Msh2-Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair. J Biol Chem 2018;293:18055-70. [Crossref] [PubMed]
- Zhao L. Mismatch repair protein expression in patients with stage II and III sporadic colorectal cancer. Oncol Lett 2018;15:8053-61. [PubMed]
- Velázquez C, Esteban-Cardenosa EM, Lastra E, et al. Unraveling the molecular effect of a rare missense mutation in BRIP1 associated with inherited breast cancer. Mol Carcinog 2019;58:156-60. [Crossref] [PubMed]
- Aravind Kumar M, Naushad SM, Narasimgu N, et al. Whole exome sequencing of breast cancer (TNBC) cases from India: association of MSH6 and BRIP1 variants with TNBC risk and oxidative DNA damage. Mol Biol Rep 2018;45:1413-9. [Crossref] [PubMed]


