
Key genes involved in cell cycle arrest and DNA damage repair identified in anaplastic thyroid carcinoma using integrated bioinformatics analysis
Introduction
Thyroid carcinoma (THCA) comprises numerous subtypes, including papillary thyroid cancer (PTC) and anaplastic thyroid carcinoma (ATC) (1). PTC has a good prognosis and is the most common subtype of THCA. In contrast, ATC is a rare subtype of THCA. However, as this subtype is therapy resistant and exhibits rapid progression, ATC has a poor prognosis (1). Currently, downregulation of breast cancer metastasis suppressor 1 (BRMS1) is a poor prognosis biomarker of ATC. Decreased BRMS1 expression promotes the proliferation and migration of cancer cells through CX43 and P53. However, further studies are still needed to identify more key genes and the underlying mechanism of ATC (2). Therefore, in this study, we aim to identify the key pathways and key genes in ATC using integrated bioinformatics methods.
Gene chips are powerful tools for identifying differentially expressed genes (DEGs). Gene chips are widely used to produce and store a significant amount of data in public databases (3). Furthermore, some bioinformatics studies on ATC have been performed (4,5), suggesting that integrated bioinformatics methods could contribute to further studies on the underlying mechanisms of ATC progression. Therefore, in this study, we aimed to identify the key pathways and key genes in ATC using integrated bioinformatics methods.
In present study, we used GSE29265 and GSE33630 from the Gene Expression Omnibus (GEO). Next, using the GEO2R online tool and Venn diagram software, common DEGs were detected in the two datasets noted above. Third, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was utilized to analyze the involvement of these DEGs in molecular function (MF), cellular component (CC), biological process (BP) and Kyoto Encyclopedia of Gene and Genome (KEGG) pathways. Fourth, we constructed protein-protein interaction (PPI) networks and then utilized Cytotype MCODE (Molecular Complex Detection) for further analysis of the DEGs to identify core genes. Then, the Kaplan-Meier (KM) plotter online database was used to search for prognostic information on these core genes in THCA patients (P<0.05). We found that ANLN, DEPDC1, KIF2C, CENPN, TACC3 CCNB2 and CDC6 might be prognostic biomarkers of THCA. We reanalyzed 54 core DEGs for KEGG pathway enrichment. Four DEGs (CHEK1, CDK1, CCNB1 and CCNB2) were generated and significantly enriched in the cell cycle pathway, especially in the G2/M phase (P<0.01). In addition, these genes were significantly enriched in the P53 pathway, especially before G2 phase arrest (P<0.01). Utilizing Gene Expression Profiling Interactive Analysis (GEPIA), we found that these four genes (CHEK1, CDK1, CCNB1 and CCNB2) were significantly associated with ANLN, DEPDC1, KIF2C, CENPN, TACC3 CCNB2 and CDC6, which might be prognostic biomarkers of THCA. We identified these four DEGs (CHEK1, CDK1, CCNB1 and CCNB2) as key genes involved in ATC.
Methods
Microarray data information
The NCBI-GEO is a powerful database that can be utilized for microarray/gene profile analysis. The gene expression profiles of ATC, PTC and normal thyroid tissues were acquired from GSE29265 and GSE33630. Both microarray datasets of GSE29265 and GSE33630 were included in the GPL570 platform [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array]. The GSE29265 dataset included 10 ATC tissues, 49 PTC tissues and 45 normal thyroid tissues, whereas the GSE33630 dataset included 9 ATC tissues, 10 PTC tissues and 10 normal thyroid tissues.
Data processing of DEGs
Using GEO2R online tools (6), DEGs between ATC specimens and normal thyroid tissue specimens and DEGs between ATC specimens and PTC specimens were identified based on an |logFC| >1 and an adjusted P value <0.05. DEGs with a logFC <0 were considered downregulated genes, while DEGs with a logFC >0 were considered upregulated genes. Then, the raw data from the two datasets in TXT format were imported into Venn software online to identify DEGs common to the two datasets.
Gene ontology (GO) and pathway enrichment analyses
Utilizing GO analysis, the characteristic biological attributes of the DEGs were identified. The functional attributes of the DEGs were identified by KEGG pathway enrichment analysis. DAVID and KEGG were employed for GO and pathway enrichment analysis (7). DAVID was used to visualize DEG enrichment in BP, MF, CC and pathways (P<0.05) (7).
PPI network construction and module analysis
We used STRING (Search Tool for the Retrieval of Interacting Genes), which is a powerful online tool, to evaluate and establish PPI networks (8). Then, we applied the STRING application in Cytoscape to detect the potential correlations between these DEGs (maximum number of interactions =0 and confidence score ≥0.4) (9). In addition, the MCODE application in Cytoscape was utilized to examine the modules of the PPI network and identify the central genes among the DEGs (degree cutoff =2, max. depth =100, k-core =2, and node score cutoff =0.2).
Survival analysis and RNA sequencing of core genes
KM plotter is a powerful online tool that assesses the effect of numerous genes on the prognosis of various tumor types based on data from the European Genome-phenome Archive, The Cancer Genome Atlas (TCGA) database and the GEO (Affymetrix microarrays only) (https://kmplot.com/analysis/index.php?p=service&cancer=pancancer_rnaseq) (10). The log rank P value and hazard ratio with 95% confidence intervals were analyzed and displayed on the plot.
Utilizing Pearson analysis of GEPIA, we analyzed the correlation between the expression levels of genes that were significantly enriched in a pathway and the expression levels of genes that were verified to be associated with the prognosis of THCA (11).
Drug relations of core genes
The DrugBank database is a comprehensive, freely accessible, online database containing information on drugs and drug targets. As both a bioinformatics and a cheminformatics resource, DrugBank combines detailed drug data with comprehensive drug target information (12). The database was used to search the drug relations of core DEGs (https://www.drugbank.ca/).
Results
Identification of DEGs in ATCs
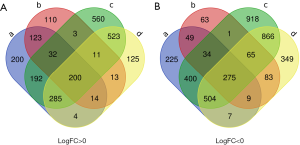
The present study utilized 19 ATC tissues, 59 PTC tissues and 55 normal thyroid tissues. Using GEO2R online tools, we extracted 3,638 and 8,202 DEGs from GSE29265 and GSE33630, respectively. Then, we applied Venn diagram software to identify DEGs common to the two datasets. A total of 475 common DEGs were detected, including 275 downregulated genes (logFC <0) and 200 upregulated genes (logFC >0) in the ATC tissues (Table 1 and Figure 1).
Table 1
| DEGS | Gene name |
|---|---|
| Up-regulated | CLMP, ITGA5, IRAK1, TPX2, GPSM2, TRIB3, IGF2BP3, CCNB1, GINS1, KPNA2, ANLN, BIRC5, MKNK1, FOXM1, CDK1, ABHD2, PXDN, HINT3, PLD1, CDC6, KLHL7, GNB4, AURKA, KIF14, NEK6, C6orf62, POC1A, MRTO4, CENPL, SIX2, KIF4A, LRR1, MIR3658/UCK2, LOC100507855/AK4, SQLE, MCM10, FAM72A/FAM72D/FAM72B/FAM72C, MELK, HN1, PARVB, MME, OIP5, CCNA2, GTSE1, E2F7, NUF2, PSRC1, CDCA5, STMN1, SLC2A1, CRNDE, CKS2, DEPDC1, KIF23, NDE1, IQGAP3, DIAPH3, FAM126A, MTFR2, CENPE, ANGPTL4, LINC00673/LINC00511, TACC3, PAX6, GALNT6, HOXD10, CCNB2, PRC1, STAB1, SRD5A1, CENPW, CEP55, CEMIP, CTPS1, ADORA3, MOCOS, SLC6A8, CDC45, EIF4EBP1, NTNG1, ADORA2B, DLGAP5, ENO1, MKI67, CDCA2, ITCH, ADM, CCBE1, COLGALT1, SHOX2, KIAA0101, HOXB7, CTTN, PROCR, SLC16A1, NEK2, COL12A1, SPC24, UGCG, LPP, DEPDC7, NETO2, APCDD1L, P3H1, LGALS1, STEAP3, CHEK1, KIF11, CENPH, KIF18B, MIR1908/FADS1, FKBP14, CDC25C, LINC01116, DCLRE1B, ZWILCH, ENAH, DBF4, NRAS, PTGFR, KIAA1549L, ASPH, KIAA1524, SATB2, INF2, GAS2L3, KIF2C, PYCR1, CDC20, CENPN, SLC4A7, GART, BUB1, PBK, SOAT1, ATL3, PRR11, TRIP13, ADA, DUXAP10, SKA3, NCAPH, TMEM158, SHCBP1, TUBB2A, MARCKS, SACS, ASPM, CDCA3, LOXL2, KNL1, PYGL, SLC7A5, STIL, GINS4, LOC654342/LOC645166, RALA, AUNIP, RRM2, ELOVL6, SLC35F6/CENPA, TOP2A, FANCI, RAD54L, FGFR1OP2, SPC25, ANPEP, KIF15, BUB1B, MTCL1, HJURP, FAM83D, RAD51, ANGPTL2, HMMR, APCDD1L-AS1, CXCL5, LINC00537, KIF20A, ORC6, FOXD1, YKT6, MAP4K4, HOXC6, UHRF1, SGO2, C18orf54, CSF1R, NPL, TTK, CDKN3, HEATR3, NCAPG, NDC1, GOLT1B, NPC1, FAM64A, CENPF, NUSAP1, CDCA8 |
| Down-regulated | FARP1, BTG2, FAM214A, CYP39A1, NAP1L2, EPB41L4B, KLHL14, PAIP1, PAPOLA, ZNF582-AS1, SORL1, NFIA, TEK, ATL2, LOC100506098, ALDH3A2, DNAJC15, BORCS7, MAOA, ADGRF1, N4BP2L2, TRIM2, UBR3, ENOSF1, CDS1, ALAD, ASS1, BDH2, MYO5C, METTL21A, SMIM10L2B/SMIM10L2A, EPHA4, NEBL, ZBTB20, HDHD2, MGST1, ASAH1, MUC15, JADE1, OLFML2A, PPL, RALGPS1, SEC63, CPVL, ARHGEF26, SVIP, MAP7, C1orf115, SH3BGRL2, CD200, PDCD6IP, MIR6883/PER1, LOC101930404/SNORD116-28/SNORD115-26/SNORD115-13/SNORD115-7/SNORD116-22/SNORD116-4/PWARSN/SNORD107/SNRPN/IPW, ESR1, KIAA0232, MIR29C/MIR29B2, FBXO3, CUL3, CLIC5, NPNT, AIF1L, SLC5A3, NELL2, MIR4680/PDCD4, NKX2-1, AUH, ALDH5A1, SSBP2, ZNF75D, ANGPTL1, KLHDC1, GLCCI1, ADGRG1, EMCN, LMO7, TSPAN12, SHE, HHEX, NTRK2, OCLN, CLDN3, SFTA3, SNX5, BBOF1, RNF146, FZD5, ZNF10, APTR, NAP1L5, MGAT4C, TMEM50B, TBL1XR1, PHACTR2, KLF5, PDE5A, LRRC70/IPO11, CBX7, RAI2, DIO2, FUCA1, MPP5, ID2, NME5, ADIRF, SOX6, TMEM243, FLRT3, ARG2, TM9SF3, SMIM5, PLPP3, DYNC2LI1, CAT, PTPRM, CELSR2, CFLAR, PIK3R1, SOWAHA, VSIG2, FOSB, CAV2, FAM46C, RMST, ASF1A, CPE, TMEM192, CTSO, HIBADH, ZNF91, TSHR, ARMCX4, KCNJ16, HNRNPU, BHLHE41, EPM2AIP1, C2orf40, CAAP1, HSD17B6, LNX1, ABLIM1, ZNF471, ZNF652, HOPX, ZSCAN18, EPB41L4A, ANXA1, MAATS1, ITIH5, IQCK, SYNE2, MFSD6, ZNF506, BMPR1A, ARHGEF12, SAMD12, SNURF/SNRPN, ATF3, ZNF148, TMEM30B, LMBRD1, MYO5B, RNF180, DUOX2, RCOR3, ERMP1, CCNG2, ITSN2, SCD5, MAL, ATP13A4, CASD1, RASEF, EGR1, USP9X, USP54, CYP4V2, DNALI1, TM7SF2, ABCA8, CDC37L1, RALGAPA2, LRIG3, KIAA1217, EGR2, FGFR2, PKP4, SLC6A13, GALNT12, DAPK2, TXNL1, CD9, TRIB1, CRYGN, PER3, BEX4, MAP3K1, PDE1A, EPHX2, KANSL1L, TPD52, RGS5, PDGFRL, MIR6778/SHMT1, MYLIP, SCAPER, MIR4738/H3F3B/H3F3A, PTPRN2, AFAP1L2, SDPR, IQCA1, UBL3, ANOS1, REEP5, HNRNPDL, CDH1, FOS, PEX3, CLU, TMEM245, ARAP2, ATP8A1, ABAT, PCM1, RBM47, SLC20A2, APLP2, LINC00271, IL6ST, SORD, INMT, CYLD, ARHGAP44, EIF4E3, 1-Mar, CADM1, TCERG1L, ANKRD12, LMO3, PPFIBP2, ZMYND11, C6orf132, CTNNAL1, SNX1, NEDD9, ZNF431, PITPNM3, PCP4, ZBED2, FRZB, CLDN8, TJP2, PAPLN, LINC01578, PPP1R14C, F11R, IFT57, DOCK3, IGIP, PHF10, NANOS1, RANBP2, KCNJ15, C1orf116, FANK1, EPB41L5, RNF128, TMED4, ZNF300P1, MAL2, CLIC6, GRAMD3, ARMCX2, NDN, EXOC6, PIK3IP1 |
GO and KEGG pathway analysis of DEGs in ATCs
All 475 DEGs were analyzed by DAVID software. The GO analysis results revealed the following: (I) for BP, upregulated DEGs were particularly enriched in regulation of cell division, mitotic nuclear division, sister chromatid cohesion, chromosome segregation, mitotic spindle organization and G2/M transition of mitotic cell cycle, and downregulated DEGs were particularly enriched in negative regulation of transcription from RNA polymerase II promoter, actomyosin structure organization, skeletal muscle cell differentiation, response to drug, positive regulation of fat cell differentiation, and neurotransmitter catabolic process. (II) For CC, upregulated DEGs were significantly enriched in the condensed chromosome kinetochore, midbody, chromosome, centromeric region, spindle pole, spindle, and kinetochore, and downregulated DEGs were particularly enriched in extrinsic component of membrane, extracellular exosome, bicellular tight junction, endoplasmic reticulum, focal adhesion, and cell-cell junction. (III) For MF, upregulated DEGs were enriched in microtubule binding, protein binding, microtubule motor activity, ATP binding, protein kinase binding and protein kinase activity, and downregulated DEGs were particularly enriched in protein homodimerization activity, cytoskeletal protein binding, RNA polymerase II core promoter proximal region sequence-specific DNA binding, zinc ion binding, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, transcriptional repressor activity, and RNA polymerase II core promoter proximal region sequence-specific binding (P<0.05, Table 2).
Table 2
| Expression | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_DIRECT | GO:0051301~cell division | 40 | 20.73 | 1.2E−28 | 1.86E−25 |
| GOTERM_BP_DIRECT | GO:0000278~mitotic nuclear division | 35 | 18.13 | 4.9E−28 | 7.74E−25 | |
| GOTERM_BP_DIRECT | GO:0007062~sister chromatid cohesion | 19 | 9.84 | 3.5E−17 | 5.60E−14 | |
| GOTERM_BP_DIRECT | GO:0007059~chromosome segregation | 15 | 7.77 | 1.2E−14 | 1.89E−11 | |
| GOTERM_BP_DIRECT | GO:0007052~mitotic spindle organization | 8 | 4.15 | 2.4E−8 | 3.81E−05 | |
| GOTERM_BP_DIRECT | GO:0000086~G2/M transition of mitotic cell cycle | 13 | 6.74 | 2.77E−8 | 4.41E−05 | |
| GOTERM_CC_DIRECT | GO:0000777~condensed chromosome kinetochore | 17 | 8.81 | 4.37E−16 | 5.66E−13 | |
| GOTERM_CC_DIRECT | GO:0030496~midbody | 19 | 9.84 | 1.09E−15 | 1.41E−12 | |
| GOTERM_CC_DIRECT | GO:0000775~chromosome, centromeric region | 14 | 7.25 | 1.42E−14 | 1.78E−11 | |
| GOTERM_CC_DIRECT | GO:0000922~spindle pole | 15 | 7.77 | 5.60E−12 | 7.08E−09 | |
| GOTERM_CC_DIRECT | GO:0005819~spindle | 14 | 7.25 | 3.21E−10 | 4.07E−07 | |
| GOTERM_CC_DIRECT | GO:0000776~kinetochore | 12 | 6.22 | 6.06E−10 | 7.67E−07 | |
| GOTERM_MF_DIRECT | GO:0008017~microtubule binding | 16 | 8.29 | 5.63E−09 | 7.54E−06 | |
| GOTERM_MF_DIRECT | GO:0005515~protein binding | 130 | 67.36 | 1.73E−08 | 2.32E−05 | |
| GOTERM_MF_DIRECT | GO:0003777~microtubule motor activity | 9 | 4.66 | 1.98E−06 | 0.002653 | |
| GOTERM_MF_DIRECT | GO:0005524~ATP binding | 34 | 17.62 | 3.53E−05 | 0.047295 | |
| GOTERM_MF_DIRECT | GO:0019901~protein kinase binding | 15 | 7.77 | 4.65E−05 | 0.062196 | |
| GOTERM_MF_DIRECT | GO:0004672~protein kinase activity | 13 | 6.74 | 4.37E−04 | 0.583283 | |
| Down-regulated | GOTERM_BP_DIRECT | GO:0000122~negative regulation of transcription from RNA polymerase II promoter | 25 | 9.43 | 2.26E−05 | 0.037204 |
| GOTERM_BP_DIRECT | GO:0031032~actomyosin structure organization | 5 | 1.89 | 3.84E−04 | 0.628799 | |
| GOTERM_BP_DIRECT | GO:0035914~skeletal muscle cell differentiation | 6 | 2.26 | 4.22E−04 | 0.691557 | |
| GOTERM_BP_DIRECT | GO:0042493~response to drug | 12 | 4.53 | 0.002132 | 3.447900 | |
| GOTERM_BP_DIRECT | GO:0045600~positive regulation of fat cell differentiation | 5 | 1.89 | 0.003185 | 5.108681 | |
| GOTERM_BP_DIRECT | GO:0042135~neurotransmitter catabolic process | 3 | 1.13 | 0.004462 | 7.088059 | |
| GOTERM_CC_DIRECT | GO:0019898~extrinsic component of membrane | 8 | 3.02 | 1.09E−04 | 0.141680 | |
| GOTERM_CC_DIRECT | GO:0070062~extracellular exosome | 58 | 21.89 | 5.59E−04 | 0.724929 | |
| GOTERM_CC_DIRECT | GO:0005923~bicellular tight junction | 8 | 3.02 | 7.82E−04 | 1.013103 | |
| GOTERM_CC_DIRECT | GO:0005783~endoplasmic reticulum | 23 | 8.68 | 0.001588 | 2.047973 | |
| GOTERM_CC_DIRECT | GO:0005925~focal adhesion | 13 | 4.91 | 0.006215 | 7.798362 | |
| GOTERM_CC_DIRECT | GO:0005911~cell-cell junction | 8 | 3.02 | 0.008202 | 10.170454 | |
| GOTERM_MF_DIRECT | GO:0042803~protein homodimerization activity | 23 | 8.68 | 2.96E−04 | 0.409611 | |
| GOTERM_MF_DIRECT | GO:0008092~cytoskeletal protein binding | 6 | 2.26 | 4.23E−04 | 0.586252 | |
| GOTERM_MF_DIRECT | GO:0000978~RNA polymerase II core promoter proximal region sequence-specific DNA binding | 14 | 5.28 | 9.54E−04 | 1.317253 | |
| GOTERM_MF_DIRECT | GO:0008270~zinc ion binding | 27 | 10.19 | 0.006690 | 8.900309 | |
| GOTERM_MF_DIRECT | GO:0001077~transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 9 | 3.40 | 0.013679 | 17.407838 | |
| GOTERM_MF_DIRECT | GO:0001078~transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding | 6 | 2.26 | 0.016213 | 20.306251 |
BP, biological processes; MF, molecular function; CC, cell component; GO, gene ontology.
KEGG analysis results suggested that upregulated DEGs were particularly enriched in the Cell cycle, P53 signaling pathway, Oocyte meiosis, Progesterone-mediated oocyte maturation and MicroRNAs in cancer and downregulated DEGs were particularly enriched in Tight junction, Cell adhesion molecules, Tryptophan metabolism, Valine, leucine and isoleucine degradation, and Signaling pathways regulating pluripotency of stem cells (P<0.05, Table 3). CCNB1, CDK1, CCNB2 and CHEK1 were markedly enriched in the cell cycle pathway and p53 signaling pathway (P<0.01, Table 3).
Table 3
| Expression | Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|---|
| Up-regulated | hsa04110 | Cell cycle | 14 | 7.25 | 7.79E−10 | CCNB1, CDK1, CCNB2, CHEK1, CDC25C, BUB1, CCNA2, CDC45, CDC6, DBF4, TTK, BUB1B, ORC6, CDC20 |
| hsa04115 | p53 signaling pathway | 7 | 3.63 | 9.38E−05 | CCNB1, CDK1, CCNB2, CHEK1, STEAP3, RRM2, GTSE1 | |
| hsa04114 | Oocyte meiosis | 7 | 3.63 | 0.001442 | CCNB1, CDK1, CCNB2, CDC25C, BUB1, AURKA, CDC20 | |
| hsa04914 | Progesterone-mediated oocyte maturation | 6 | 3.11 | 0.002787 | CCNB1, CDK1, CCNB2, CDC25C, BUB1, CCNA2 | |
| hsa05206 | MicroRNAs in cancer | 8 | 4.15 | 0.040192 | CDC25C, KIF23, NRAS, ITGA5, MARCKS, STMN1, CDCA5, HOXD10 | |
| Down-regulated | hsa04530 | Tight junction | 6 | 2.26 | 0.005985 | CLDN8, F11R, OCLN, CLDN3, MPP5, TJP2 |
| hsa04514 | Cell adhesion molecules (CAMs) | 7 | 2.64 | 0.011608 | CLDN8, F11R, OCLN, PTPRM, CADM1, CLDN3, CDH1 | |
| hsa00380 | Tryptophan metabolism | 4 | 1.51 | 0.016006 | MAOA, CAT, ALDH3A2, INMT | |
| hsa00280 | Valine, leucine and isoleucine degradation | 4 | 1.51 | 0.024573 | ABAT, ALDH3A2, HIBADH, AUH | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 6 | 2.26 | 0.039046 | FGFR2, ID2, IL6ST, FZD5, PIK3R1, BMPR1A |
PPI network and modular analysis
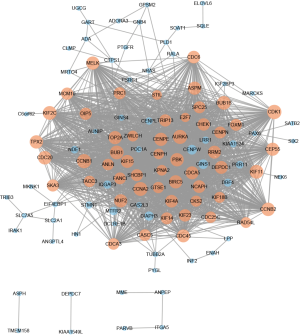
A total of 116 DEGs were imported into the DEG PPI network complex, which included 116 nodes and 1,741 edges. The 116 nodes were all upregulated genes (Figure 2). In total, 359 of the 475 DEGs were not included in the PPI network (Figure 2). Then, we utilized Cytotype MCODE for further analysis. In total, 54 central nodes were all upregulated genes, and these central nodes were identified among the 116 nodes (Figure 2).
Reanalysis of 54 selected genes by KEGG pathway enrichment
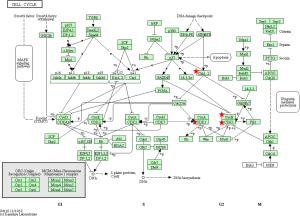
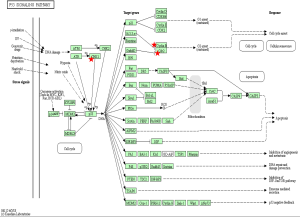
To identify the pathways associated with the 54 core DEGs, these 54 genes were reanalyzed by KEGG pathway enrichment through DAVID (P<0.05, Table 4). Four genes (CCNB1, CCNB2, CDK1 and CHEK1) were significantly enriched in the Cell cycle pathway, especially in the G2/M phase (P<0.01 Table 4 and Figure 3). In addition, these genes were significantly enriched in the P53 pathway, especially before G2 phase arrest (P<0.01, Table 4 and Figure 4).
Table 4
| Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|
| hsa04110 | Cell cycle | 11 | 20.75 | 7.50E−13 | CCNB1, CDK1, CCNB2, CHEK1, CDC45, CDC6, BUB1, BUB1B, CDC20, CDC25C, CCNA2 |
| hsa04114 | Oocyte meiosis | 7 | 13.21 | 6.87E−07 | CCNB1, CDK1, CCNB2, BUB1, AURKA, CDC20, CDC25C |
| hsa04115 | p53 signaling pathway | 6 | 11.32 | 1.36E−06 | CCNB1, CDK1, CCNB2, CHEK1, RRM2, GTSE1 |
| hsa04914 | Progesterone-mediated oocyte maturation | 6 | 11.32 | 5.00E−06 | CCNB1, CDK1, CCNB2, BUB1, CDC25C, CCNA2 |
| hsa05203 | Viral carcinogenesis | 4 | 7.55 | 0.023352 | CDK1, CHEK1, CDC20, CCNA2 |
Analysis of core genes by the KM plotter and GEPIA
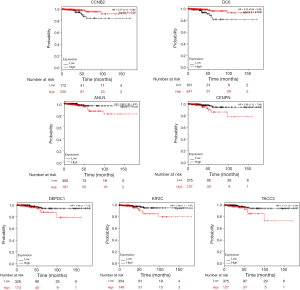
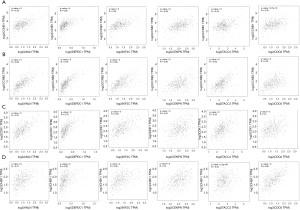
We utilized the KM plotter to analyze the survival data of the 54 core genes. Five genes with high expression (ANLN, DEPDC1, KIF2C, CENPN and TACC3) were associated with significantly worse survival, whereas 2 genes (CCNB2 and CDC6) were associated with significantly better survival (P<0.05, Figure 5). Using GEPIA, we found that CCNB1, CDK1, CCNB2 and CHEK1 expression was significantly positively correlated with the expression of ANLN, DEPDC1, KIF2C, CENPN, TACC3 and CDC6. (P<0.01, Figure 6).
Drug relations of core genes
Using the online tool, CHEK1, CDK1, and TOP2A were found in the DrugBank database. Fostamatinib is an inhibitor of CHEK1 and CDK1. In total, 20 approved drugs were identified as inhibitors of TOP2A. However, four drugs identified as inhibitors of TOP2A were classified as investigational, withdrawn or experimental (Table 5).
Table 5
| Gene name | No. | Drug bank ID | Name | Drug group | Pharmacological action | Actions |
|---|---|---|---|---|---|---|
| CHEK1 | 1 | DB12010 | Fostamatinib | Approved, investigational | Unknown | Inhibitor |
| CDK1 | 1 | DB12010 | Fostamatinib | Approved, investigational | Unknown | Inhibitor |
| 2 | DB04014 | Alsterpaullone | Experimental | Unknown | Inhibitor | |
| 3 | DB02116 | Olomoucine | Experimental | Unknown | Binder | |
| 4 | DB02052 | Indirubin-3'-monoxime | Experimental | Unknown | Binder | |
| 5 | DB03428 | SU9516 | Experimental | Unknown | Binder | |
| TOP2A | 1 | DB00218 | Moxifloxacin | Approved, investigational | Unknown | Inhibitor |
| 2 | DB00276 | Amsacrine | Approved, investigational | Yes | Inhibitor | |
| 3 | DB00380 | Dexrazoxane | Approved, withdrawn | Yes | Inhibitor | |
| 4 | DB00385 | Valrubicin | Approved | Yes | Inhibitor | |
| 5 | DB00444 | Teniposide | Approved | Yes | Inhibitor | |
| 6 | DB00445 | Epirubicin | Approved | Unknown | Inhibitor | |
| 7 | DB00467 | Enoxacin | Approved, investigational | No | Inhibitor | |
| 8 | DB00487 | Pefloxacin | Approved | Unknown | Inhibitor | |
| 9 | DB00694 | Daunorubicin | Approved | Unknown | Inhibitor | |
| 10 | DB00773 | Etoposide | Approved | Unknown | Inhibitor | |
| 11 | DB00970 | Dactinomycin | Approved, investigational | Yes | Inhibitor | |
| 12 | DB00978 | Lomefloxacin | Approved, investigational | Yes | Inhibitor | |
| 13 | DB00997 | Doxorubicin | Approved, investigational | Unknown | Inhibitor | |
| 14 | DB01059 | Norfloxacin | Approved | Unknown | Inhibitor | |
| 15 | DB01137 | Levofloxacin | Approved, investigational | Yes | Inhibitor | |
| 16 | DB01165 | Ofloxacin | Approved | Unknown | Inhibitor | |
| 17 | DB01177 | Idarubicin | Approved | Yes | Inhibitor | |
| 18 | DB01179 | Podofilox | Approved | Yes | Inhibitor | |
| 19 | DB01204 | Mitoxantrone | Approved, investigational | Yes | Inhibitor | |
| 20 | DB01208 | Sparfloxacin | Approved, investigational, withdrawn | Yes | Inhibitor | |
| 21 | DB04576 | Fleroxacin | Experimental | Yes | Inhibitor | |
| 22 | DB04967 | Lucanthone | Investigational | Yes | Inhibitor | |
| 23 | DB06013 | Aldoxorubicin | Investigational | Yes | Inhibitor | |
| 24 | DB09047 | Finafloxacin | Approved, investigational | Yes | Inhibitor |
Discussion
In this study, we identified important genes and pathways in ATCs using bioinformatics methods. DEGs between ATC specimens and normal thyroid tissue specimens as well as DEGs between ATC specimens and PTC specimens were identified from two original microarray datasets using GEO2R software. Then, the DEGs were integrated via Venn software. In total, 475 common DEGs were identified (Table 1).
GO analysis using DAVID methods revealed the following: (I) for BP, upregulated DEGs were particularly enriched in regulation of cell division, mitotic nuclear division, sister chromatid cohesion, chromosome segregation, mitotic spindle organization and G2/M transition of mitotic cell cycle, and downregulated DEGs were particularly enriched in negative regulation of transcription from RNA polymerase II promoter, actomyosin structure organization, skeletal muscle cell differentiation, response to drug, positive regulation of fat cell differentiation, and neurotransmitter catabolic process. (II) For CC, upregulated DEGs were significantly enriched in the condensed chromosome kinetochore, midbody, chromosome, centromeric region, spindle pole, spindle, and kinetochore, and downregulated DEGs were particularly enriched in extrinsic component of membrane, extracellular exosome, bicellular tight junction, endoplasmic reticulum, focal adhesion, and cell-cell junction. (III) For MF, upregulated DEGs were enriched in microtubule binding, protein binding, microtubule motor activity, ATP binding, protein kinase binding and protein kinase activity, and downregulated DEGs were particularly enriched in protein homodimerization activity, cytoskeletal protein binding, RNA polymerase II core promoter proximal region sequence-specific DNA binding, zinc ion binding, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, transcriptional repressor activity, and RNA polymerase II core promoter proximal region sequence-specific binding (P<0.05, Table 2). In pathway analysis, upregulated DEGs were particularly enriched in Cell cycle, P53 signaling pathway and MicroRNAs in cancer, while downregulated DEGs were particularly enriched in Tight junction, Cell adhesion molecules, Tryptophan metabolism, Valine, leucine and isoleucine degradation, and Signaling pathways regulating pluripotency of stem cells. Four upregulated DEGs (CCNB1, CDK1, CCNB2 and CHEK1) were markedly enriched in the cell cycle pathway and p53 signaling pathway (P<0.01, Table 3).
Next, a DEG PPI network complex with 116 nodes and 1,741 edges was constructed via the STRING online database and Cytoscape software. Then, 54 vital upregulated genes were screened from the PPI network complex by Cytoscape MCODE analysis. Furthermore, KM plotter analysis identified 5 genes with high expression (ANLN, DEPDC1, KIF2C, CENPN, and TACC3) that were associated with significantly worse survival (P<0.05, Figure 5). Two genes (CCNB2 and CDC6) were associated with improved survival (P<0.05, Figure 5).
Although the KM plotter database used for survival analysis includes 502 THCA patients with available clinical data and different histological subtypes, we hypothesize that ANLN, DEPDC1, KIF2C, CENPN, TACC3 CCNB2 and CDC6 may be clinically relevant genes for ATC. Further studies with larger sample sizes will be needed to identify the correlations between ANLN, DEPDC1, KIF2C, CENPN, TACC3 CCNB2 and CDC6 expression and ATC survival.
Furthermore, we reanalyzed 54 genes via DAVID for KEGG pathway enrichment and found that four genes (CCNB1, CDK1, CCNB2 and CHEK1) were markedly enriched in the cell cycle pathway (especially in the S/G2 phase) and p53 signaling pathway (especially before G2 phase arrest) (P<0.01, Table 4 and Figures 3 and 4). This finding is consistent with the results of the KEGG pathway analysis of 200 upregulated DEGs described above.
Additionally, utilizing online tool analysis of the 54 genes, CHEK1, CDK1 and TOP2A were found in the DrugBank database. Fostamatinib was verified to be an inhibitor of CHEK1 and CDK1. Meanwhile, 24 drugs were identified as inhibitors of TOP2A.
When DNA damage occurs during the G2 phase, cyclin-dependent kinase 1 (CDK1) is phosphorylated and subsequently inhibited. Protein kinases Chk1 and Chk2 (referred to as CHEK1 and CHEK2, respectively) are activated in an ATM-dependent manner (13,14). Cdc25 is phosphorylated and inactivated and subsequently binds to the CDK1-cyclin B complex (Figure 3) (13,14). The CDK1-cyclin B complex is inactivated, and the complex is sequestered from the nucleus. Thus, CDK1 inhibition and sequestration of the CDK1-cyclin B complex from the nucleus prevent cells from entering the mitotic phase and cause cell cycle arrest (Figure 3) (13,14). In the P53 pathway, CHK1 activation promotes P53 expression, which increases the transcription of p21, Gadd45 and 14-3-3 sigma (14-3-3 s). Cyclin B binds to 14-3-3 s, and the Cdc2-cyclin B complex is sequestered from the nucleus and inactive (14,15). Gadd45 binds to the Cdc2-cyclin B complex, inactivating the complex. In addition, Gadd45 and 14-3-3 bind to the Cdc2-cyclin B complex, causing G2 arrest. Furthermore, p21 binds to the cyclinD-CDK4/6 complex and cyclinE-CDK2 complex. The two complexes are inhibited, and the cell cycle is arrested in G1 (Figure 4) (14,15).
CHEK1 regulates key genes involved in cell cycle arrest and DNA repair in the cell cycle pathway and P53 pathway. Therefore, CHEK1 (also known as Chk1) is a central gene of the cell cycle that regulates cell cycle checkpoints to prevent cells with damaged DNA from undergoing mitosis and promotes various aspects of DNA repair.
Previous studies found that CHEK1 overexpression was associated with poor outcomes of NSCLC and ovarian cancer. CHEK1 is a potential target for tumor therapy (16,17). Numerous studies have shown that the efficacy of radiotherapy and chemotherapy increase when administered in combination with CHEK1 inhibitors in several cancers (18-20). Clinical trials are ongoing to test selective CHK1 inhibitors in cancer patients. Several studies have found that CHK1 inhibitors enhance the efficacy of multiple DNA-damaging therapies. Given its role in regulating cell cycle checkpoints, CHK1 is regarded as a useful target for cancer therapy. Numerous studies are being conducted to develop CHK1 inhibitors as single-agent therapies (21-24).
CDK1 is a key gene that promotes cell entry into and progression through mitosis (14). When DNA damage occurs in the G2 phase checkpoint, the WEE1 and MYT1 kinases inhibit CDK1 through phosphorylation. Phosphorylated CDK1 is inactivated and down-regulated. Thus, Cdc25, which can phosphorylate and reactive CDK1, is inactive (Figure 3) (25). Previous studies showed that CDK1 was necessary for DNA replication and prevention of replication-associated DNA damage. Via phosphorylation of multiple downstream targets, CDK1 promotes replicative DNA synthesis (26). Since CDK1 plays a critical role in the checkpoint, it is hypothesized that CDK1 inhibitors will enhance the killing effects of replication-toxic agents in cancer cells (26). Furthermore, numerous studies have shown that CDK1 inhibitors inhibit proliferation of lung, colon and pancreatic cancer cell lines or induced tumor cell apoptosis (14).
Fostamatinib is an inhibitor of CDK1 and CHEK1 and is approved by the United States Food and Drug Administration for the treatment of thrombocytopenia in adult patients with chronic immune thrombocytopenia (27,28). The drug is also being assessed for potential indications, such as chronic lymphocytic leukemia and solid tumors (27). However, the efficacy of fostamatinib as a CDK1 and CHEK1 inhibitor in the treatment of ATC requires further studies.
CCNB1 (also known as CyclinB1) is significantly overexpressed in various cancer types (29). CCNB1 binds to Cdc2 to form a maturation-promoting factor that plays key roles in the checkpoint of the cell cycle pathway in G2/M phase arrest (Figures 3 and 4) (30). Using GEPIA (11), the gene expression profile across tumor samples and paired normal tissues showed that CCNB1 is expressed in numerous tumor types and normal tissues (http://gepia.cancer-pku.cn/detail.php?gene=CCNB1). The analysis reveals significantly different expression in multiple tumor types and the corresponding normal tissue. These findings suggest that CCNB1 plays an important role in tumor transformation and progression. Previous studies showed that CCNB1 is a powerful predictive biomarker for distant metastasis-free survival, disease-free survival, recurrence-free survival and overall survival of ER+ breast cancer patients (29). Thus, CCNB1 might be a useful target for cancer management.
CCNB2 is a key component of the cell cycle pathway. CCNB2 binds to CDK1 to form a complex and regulates the activities of CDK1 via phosphorylation (14,31). Upon phosphorylation, the CCNB2-CDK1 complex is inactivated, and cells are arrested at the G2/M transition (Figures 3 and 4) (14,31). Numerous studies reported significant overexpression of CCNB2 in multiple tumors, including breast cancer, adrenocortical carcinoma, colorectal carcinoma, and gastric carcinoma (32-35). A previous study found that the expression of circulating CCNB2 mRNA in lung cancer patients was significantly higher than that in normal controls and patients with benign diseases. CCNB2 mRNA expression was significantly correlated with cancer stage and metastasis status (36). Furthermore, a previous study found that high expression of CCNB2 was an independent unfavorable prognostic factor for the overall survival of non-small-cell lung cancer patients (37). However, our study found that THCA patients with higher expression of CCNB2 exhibited better survival than patients with low CCNB2 expression. More studies on ATC should be performed in the future.
CHEK1, CDK1, CCNB1 and CCNB2 are key genes involved in cell cycle arrest and DNA damage repair in the cell cycle pathway and P53 pathway in ATC.
Five genes with high expression (ANLN, DEPDC1, KIF2C, CENPN and TACC3) were associated with significantly worse survival (Figure 5), whereas 2 genes (CCNB2 and CDC6) were associated with significantly better survival (Figure 5). However, studies on these genes are limited in THCA, especially with regard to ATC. Thus, although these genes may be potential biomarkers of ATC prognosis, further studies are required in the future.
Furthermore, CCNB1, CDK1, CCNB2 and CHEK1 expression was significantly associated with ANLN, DEPDC1, KIF2C, CENPN, TACC3 and CDC6 expression (Figure 6). Given the regulatory role of these genes in various pathways, especially the cell cycle pathway and P53 pathway, these genes (CCNB1, CDK1, CCNB2 and CHEK1) may regulate the expression of ANLN, DEPDC1, KIF2C, CENPN, TACC3 and CDC6. Via regulation of cell cycle arrest and the expression of these genes (ANLN, DEPDC1, KIF2C, CENPN, TACC3 and CDC6), CCNB1, CDK1, CCNB2 and CHEK1 may play critical roles in ATC progression and survival. Taken together, our results lead us to speculate that CCNB1, CDK1, CCNB2 and CHEK1 are key genes in ATC.
Numerous studies have found that these four genes (CCNB1, CCNB2, CDK1 and CHEK1) are related to progression in multiple tumor types. However, very few studies on these genes in ATC were found on the PubMed website. Additionally, compared with similar recently published studies that analyzed DEGs between ATC specimens and normal thyroid tissues (4,5), our study used a stricter cut-off to analyze the DEGs between ATC tissues and PTC or normal thyroid tissues. Three common DEGs were screened for further identification of potential new drugs, and survival biomarkers were also reported in our study. Therefore, the data in our study could provide useful information and directions for future studies on ATC.
Conclusions
Our bioinformatics analysis study identified four DEGs (CCNB1, CDK1, CCNB2 and CHEK1) between ATC tissues and normal thyroid tissues as well as ATC tissues and PTC tissues using different microarray datasets. Our study results indicate that since these genes are involved in cell cycle arrest and DNA repair in the cell cycle pathway and P53 pathway, these four genes might play key roles in the progression of ATC. In addition, our study highlights some clinically relevant genes that should be assessed in future studies to identify the prognosis biomarkers of ATC. However, further studies should be conducted to verify these predictions in the future.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2829). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research did not involve human participants or animals. Data were collected from public databases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haddad RI, Lydiatt WM, Ball DW, et al. Anaplastic Thyroid Carcinoma, Version 2.2015. J Natl Compr Canc Netw 2015;13:1140-50. [Crossref] [PubMed]
- Wu Y, Wang H, Zhi J, et al. BRMS1 downregulation is a poor prognostic biomarker in anaplastic thyroid carcinoma patients. Onco Targets Ther 2019;12:6937-45. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Hu S, Liao Y, Chen L. Identification of Key Pathways and Genes in Anaplastic Thyroid Carcinoma via Integrated Bioinformatics Analysis. Med Sci Monit 2018;24:6438-48. [Crossref] [PubMed]
- Gao X, Wang J, Zhang S. Integrated Bioinformatics Analysis of Hub Genes and Pathways in Anaplastic Thyroid Carcinomas. Int J Endocrinol 2019;2019:9651380.
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Author Correction: Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:11515. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Wishart DS, Feunang YD, Guo A, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074-82. [Crossref] [PubMed]
- Zeng Y, Forbes KC, Wu Z, et al. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 1998;395:507-10. [Crossref] [PubMed]
- Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 2003;36:131-49. [Crossref] [PubMed]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20:1803-15. [Crossref] [PubMed]
- Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget 2015;6:9445-56. [Crossref] [PubMed]
- Kumar G, Breen EJ, Ranganathan S. Identification of ovarian cancer associated genes using an integrated approach in a Boolean framework. BMC Syst Biol 2013;7:12. [Crossref] [PubMed]
- Mohni KN, Kavanaugh GM, David C. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res 2014;74:2835-45. [Crossref] [PubMed]
- Yashiro T, Koyama-Saegusa K, Imai T, et al. Inhibition of potential lethal damage repair and related gene expression after carbon-ion beam irradiation to human lung cancer grown in nude mice. J Radiat Res 2007;48:377-83. [Crossref] [PubMed]
- Höglund A, Nilsson LM, Muralidharan SV, et al. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin Cancer Res 2011;17:7067-79. [Crossref] [PubMed]
- McNeely S, Beckmann R, Lin AKB. CHEK again: Revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther 2014;142:1-10. [Crossref] [PubMed]
- Garrett MD, Ian C. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol Sci 2011;32:308-16. [Crossref] [PubMed]
- Carrassa L, Damia G. Unleashing Chk1 in cancer therapy. Cell Cycle 2011;10:2121-8. [Crossref] [PubMed]
- Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med 2011;17:88-96. [Crossref] [PubMed]
- Branzei D, Ciliberto A. Not all roads lead to Cdk1. Cell Cycle 2017;16:395-6. [Crossref] [PubMed]
- Liao H, Ji F, Ying S. CDK1: beyond cell cycle regulation. Aging (Albany NY) 2017;9:2465-6. [Crossref] [PubMed]
- Markham A. Fostamatinib: First Global Approval. Drugs 2018;78:959-63. [Crossref] [PubMed]
- Moore DC, Gebru T, Muslimani A. Fostamatinib for the treatment of immune thrombocytopenia in adults. Am J Health Syst Pharm 2019;76:789-94. [Crossref] [PubMed]
- Ding K, Li W, Zou Z, et al. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses 2014;83:359-64. [Crossref] [PubMed]
- Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol 2000;12:658-65. [Crossref] [PubMed]
- Petri ET, Alessia E, Lourdes E, et al. The crystal structure of human cyclin B. Cell Cycle 2007;6:1342-9. [Crossref] [PubMed]
- Fernandez-Ranvier GG, Julie W, Ru-Fang Y, et al. Identification of biomarkers of adrenocortical carcinoma using genomewide gene expression profiling. Arch Surg 2008;143:841-6. [Crossref] [PubMed]
- Shubbar E, Kovács A, Hajizadeh S, et al. Elevated cyclin B2 expression in invasive breast carcinoma is associated with unfavorable clinical outcome. BMC Cancer 2013;13:1-10. [Crossref] [PubMed]
- Park SH, Yu GR, Kim WH, et al. NF-Y-dependent cyclin B2 expression in colorectal adenocarcinoma. Clin Cancer Res 2007;13:858-67. [Crossref] [PubMed]
- Wang DG, Chen G, Wen XY, et al. Identification of biomarkers for diagnosis of gastric cancer by bioinformatics. Asian Pac J Cancer Prev 2015;16:1361-5. [Crossref] [PubMed]
- Mo ML, Chen Z, Li J, et al. Use of serum circulating CCNB2 in cancer surveillance. Int J Biol Markers 2010;25:236-42. [Crossref] [PubMed]
- Qian X, Song X, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 2015;74:222-7. [Crossref] [PubMed]


