HLA-DPB1 and Epstein-Barr virus gp42 protein jointly contribute to the development of Hodgkin lymphoma
Introduction
Despite having a high survival rate (1,2), at least 80% of patients can be cured with current chemoradiotherapy, as many as 15–20% of patients with Hodgkin lymphoma (HL) experience relapse or develop a second tumor, and in advanced patients, less than 5 percent survive without treatment. HL is most common in children and adolescents (3), and boys are 2–3 times more likely to develop this malignancy (4). However, the etiologic and pathogenic mechanisms of HL have not yet been illuminated.
Epstein-Barr virus (EBV), a spherical, 180–200 nm DNA virus, is part of the gamma-herpesviruses subfamily. Structurally, EBV comprises a genome, capsid, and envelope. As a common human virus, a significant proportion of people acquire EBV at some point in their life. With tumorigenic potential (5), EBV is associated with the pathogenesis of nasopharyngeal carcinoma, gastric cancer, Burkitt lymphoma, and HL. Although HL typically originates in B-cells, epithelial cells also play a crucial role in its pathogenesis. After B lymphocytes are infected by EBV, the host does not experience any signs or symptoms directly; rather, the virus enters a stage of latent infection, during which the B cells secrete latency markers including EBV-encoded small RNAs (EBER), latent membrane protein 1 (LMP1), and EB nuclear antigen 2 (EBNA2) (5-7). These markers stimulate an immune response from T cells, but the response is too weak to eliminate the persistent infection of EBV. Epithelial cells are jointly stimulated by interferon gamma (IFN-γ) released by T cells and the external EBV to produce HLA-II receptors and microRNA 200s protein (miR200s). miR200s can trigger B lymphocytes to transform from a state of latent infection to a state of malignant proliferation, resulting in the development of HL (Figure 1) (8).
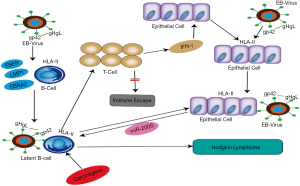
What is novel in our paper is to explore the binding mechanism between HLA-DPB1 and gp42 protein on the surface of EBV. The gp42 protein on the surface of EBV determines membrane fusion and virus entry (9,10). Gp42 and HLA-II molecules form a complex at a ratio of 1:1 (11). Gp42 also forms a high-affinity complex with gH and gL via the N-terminus (12). The binding of the C-terminus of gp42 to the β-chain of the HLA-II receptor (13) activates EBV infection of B cells and epithelial cells, leading to disease onset. HLA-DPB1 is a functional gene located on the β-chain of HLA-DP (14). In our previous study, sequence-based typing (SBT) was used to detect HLA-DPB1 genotypes in 100 HL patients and 100 healthy adults. Four HL-related genotypes were identified: HLA-DPB1*04:01:01, 04:02:01, 09:01:01, and 14:01:01. Among them, HLA-DPB1*04:01:01 had the highest expression level. In our current study, we further explored the pathogenic mechanisms of HL by studying the binding pattern between HLA-DPB1 and gp42.
Methods
Molecular dynamics simulation
The HLA-DPB1*04:01:01 genotype obtained by sequencing was further identified using BLAST in the UniProt (https://www.uniprot.org), which yielded that it was 98.4% P04440. The crystal structures of two proteins, P04440 (PDB:3LQZ) and gp42 (PDB:3FD4), were downloaded from the Protein Data Bank (PDB, http://www.rcsb.org). Molecular dynamics simulations of these two proteins (3LQZ and 3FD4) were performed using the Discovery Studio 16.1 software as follows: (I) docking simulations of these two molecules were performed using Z-DOCK (Dock Proteins); (II) the optimal docking model was screened out using R-DOCK (Refine Docked Proteins); and (III) analysis of protein interface was performed for the optimal docking molecular model.
Laboratory methods
Cell culture
First, 10 mL of fetal bovine serum (FBS) (Excell Bio, China) was added into 90 mL of IMDM medium (GIBCO, USA). Then, 1 mL of penicillin/streptomycin (GIBCO, USA) was added, and the thawed T1 cells (174 x CEM.T1, ATCC CRL-1991, USA) or 293T cells (GIBCO, USA) were put into 15-mL centrifuge tubes, respectively. After centrifugation, the supernatant was discarded, and the cells were inoculated into culture flasks (Corning Company, China) and cultured in a 5% CO2 incubator (Heal Force, Shanghai) at 37 °C with saturated humidity. The cells were passaged upon reaching 90% confluence (25 cm2).
Cell transfection
T1 cells that had grown well were treated with antibiotic-free medium and single-cell suspensions (1×105 cells/mL) were prepared. Then, the suspended cells were inoculated into 25-cm2 cell culture flasks (5 mL/vial). Three 2-mL EP tubes were labeled as A, B and C. Tube A contained 250 µL of serum-free antibiotic-free medium and 15 µL of Lipofectamine 3000 reagent (Invitrogen, China), which were gently mixed; tube B contained 250 µL of serum-free antibiotic-free medium and 15 µL of Lipofectamine 3000 reagent (Invitrogen company, China), which were gently mixed; and tube C contained 500 µL of serum-free antibiotic-free medium, 15 µg of empty plasmid pEGFP-N2 (General Biosystems, China), and 20 µL of P3000 reagent (Invitrogen, China), which were gently mixed. After culture in a 5% CO2 incubator at 37 °C with saturated humidity for 48 and 72 h, the cells were harvested. Flow cytometry assay was performed to detect the proportion of EGFP-positive cells (green fluorescence) and to identify the optimal transfection conditions. The same procedure was carried out to transfect the 293T cells. Although 293T cells are epithelial cells, they are often used as tool cells to improve transfection efficiency. Our experiments showed the transfection efficiency of T1 cell is very low. The purpose of this experiment is to verify the binding force between gp42 and HLA-DPB1 subtype, so we chose 293T cell as our experimental cells.
Plasmid construction
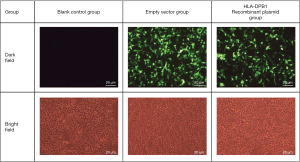
The chemically synthesized pEGFP-N2-HLA-DPB1 (General Biosystems) bacterial solution was plated on nutrient agar. First, the grown monoclonals were digested (EcoRI and BamHI), and then the positive clones were chosen for sequencing and alignment. To confirm whether the recombinant vector had been successfully constructed, polymerase chain reaction (PCR) was performed on the recombinant plasmids. These plasmids were divided into the blank control group, the empty vector group, and the HLA-DPB1 recombinant vector group.
Detection of HLA-DPB1 expression level
After the successful construction of the plasmids, flow cytometry (BD, USA) was performed to detect the proportion of EGFP-positive cells, the HLA-DPB1 gene expression level was determined by qRT-PCR (Applied Biosystems, USA), and Western blotting (Bio-Rad, USA) was used to measure the level of HLA-DPB1 protein expression.
Detection of the HLA-DPB1*gp42 binding rate
The gp42/gHgL complex protein was labeled with 6X His antibody (QIAGEN, China), after which the protein fluoresced red. Under the optimal transfection conditions, the numbers of both green- and red-fluoresced T1 cells and 293T cells under the electron microscope were counted to obtain the proportion of HLA-DPB1+ gp42+ cells; the HLA-DPB1*gp42 binding rate was calculated accordingly.
Statistical analysis
All data were expressed as mean ± standard deviation (). Statistical analysis was performed in SPSS 19.0 software package. Normally distributed data were analyzed with one-way ANOVA, with Fisher’s least significant difference (LSD) method used for unequal variances and Tamhane test used for multiple comparisons with unequal variances. A P value of 0.05 was considered to be statistically significant. The non-normal set of data was normalized by logarithm transformation, followed by one-way ANOVA.
Results
Properties of HLA-DPB1*gp42 complex
Molecular dynamics simulation using Discovery Studio 16.1 software showed that gp42 protein bound to the β-chain of HLA-DPB1 via its α-chain (Figure 2A,B) to form a complex (Figure 2C). Figure 2D shows the 3D structure of the complex. As shown in Figure 3, the gp42*HLA-DPB1 complex was a hydrophilic protein containing a total of 338 amino acids and 5,370 atoms, with a molecular weight of 39,144.12. An Instability index of 47.72 showed that the complex was unstable. Furthermore, 7 amino acids in the α-chain were bonded with 7 amino acids in the β-chain via 9 hydrogen bonds. Table 1 shows the specific amino acid names and hydrogen bond distances.
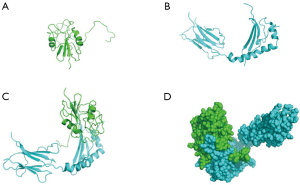
Table 1
| HLA-DPB1 residue | gp42 residue | Interaction constituents | Distance | Type |
|---|---|---|---|---|
| B:LEU-17 | A:GLU207 | B:LEU-17:HN-A:GLU207:OE1 | 2.48 | Conventional |
| B:PHE-15 | A:HIS206 | B:PHE-15:HN-A:HIS206:O | 2.0305 | Conventional |
| B:ASN6 | A:TYR167 | B:ASN6:HD21-A:TYR167:OH | 1.9158 | Conventional |
| B:ARG12 | A:GLN89 | B:ARG12:HE-A:GLN89:OE1 | 2.7927 | Conventional |
| B:ARG12 | A:GLN89 | B:ARG12:HH21-A:GLN89:OE1 | 2.664 | Conventional |
| B:ARG32 | A:PRO91 | B:ARG32:HN-A:PRO91:O | 2.7459 | Conventional |
| B:ARG32 | A:VAL90 | B:ARG32:HH11-A:VAL90:O | 2.5246 | Conventional |
| B:PRO-16 | A:HIS206 | B:PRO-16:CA-A:HIS206:O | 3.1432 | Carbon |
| B:SER3 | A:THR95 | B:SER3:CB-A:THR95:OG1 | 3.1472 | Carbon |
Optimal transfection conditions of 293T cells
After HLA-DPB1-containing plasmid vectors were constructed through transfection of T1 lymphocytes and 293T cells, flow cytometry was performed to determine the optimal transfection conditions for T1 cells. The optimal concentration of Lipofectamine 3000 was 1.5 µL/well and the optimal transfection time was 72 h; however, the positive rate was only 8.467%. The optimal transfection conditions for 293T cells were also determined by flow cytometry. The optimal concentration of the transfection reagent was 1.5 µL/well and the optimal transfection time was 48 h; under such conditions, the proportion of EGFP-positive 293T cells was 55.233% (Figure 4), which was significantly higher than that of T1 cells. As the low transfection efficiency of the T1 cells was likely to result in a high false-negative rate, the 293T cells, with their relatively high transfection efficiency, were subsequently used to detect the HLA-DPB1*gp42 binding rate. As shown in Figure 5, the quantitative results of Western blotting on 293T cells after transfection showed significant differences between the blank control group, the empty vector group, and the HLA-DPB1 recombinant vector group.

The optimal binding efficiency of HLA-DPB1 and gp42 protein
The binding efficiency of HLA-DPB1 and gp42 protein is shown in Figure 6. The binding efficiency of gp42 protein to HLA-DPB1 was highest at 80 µg (Table 2). 293T cells, which are epithelial cells derived from human embryonic kidney cells, contain plasmids containing SV40 large T antigen (Tag) origin of replication. As shown in Table 3, in the blank control group, the proportion of gp42+ cells was about 15%. Meanwhile, the proportion of HLA-DPB1+ gp42+ cells was significantly higher in the empty vector group than in the blank control group. These results suggest that 293T cells could also express proteins that can bind to gp42, among which the SV40 protein is highly probable. The proportions of gp42+ cells (%), HLA-DPB1+ cells (%), HLA-DPB1+ gp42+ cells (%) in the HLA-DPB1 recombinant plasmid group were significantly higher than those in the blank control or empty control groups, which indicates that the plasmid protein could bind to gp42. In the blank control group, the proportion of HLA-DP+ gp42+ cells was consistent with that of HLA-DPB1+ cells. In the empty vector group and HLA-DPB1 recombinant plasmid group, the proportion of HLA-DPB1+gp42+ cells was consistent with that of gp42+ cells. Therefore, the HLA-DPB1*gp42 binding efficiency in 293T cells can be obtained by calculating the proportion of gp42+ cells. In our current experiment, the HLA-DPB1*gp42 binding efficiency in 293T cells was 26–31.3%.
Table 2
| gp42 protein (μg) | HLA-DPB1+ (%) | gp42+ (%) | HLA-DP+ gp42+ (%) |
|---|---|---|---|
| 0 | 44.433±1.595 | 0.367±0.153 | 0.267±0.208 |
| 20 | 42.567±0.666 | 13.967±0.153▼ | 12.400±0.100▼ |
| 40 | 43.100±1.136 | 20.100±0.781▼△ | 18.433±0.379▼△ |
| 80 | 42.900±1.015 | 29.333±0.208▼△▲ | 27.067±0.231▼△▲ |
▼, P<0.05, compared with 0 μg; △, P<0.05, compared with 20 μg; ▲, P<0.05, compared with 40 μg.
Table 3
| Group | HLA-DPB1+ (%) | gp42+ (%) | HLA-DPB1+gp42+ (%) |
|---|---|---|---|
| Blank control group | 1.333±0.416 | 15.933±4.350 | 1.233±0.321 |
| Empty vector group | 49.900±0.300▼ | 15.767±0.451 | 15.833±0.462▼ |
| HLA-DPB1 recombinant plasmid group | 49.367±4.015▼ | 28.633±2.669▼△ | 28.900±2.951▼△ |
▼, P<0.05, compared with the blank control group; △, P<0.05, compared with the empty vector group.
Discussion
EBV can infect both epithelial cells and B lymphocytes, although it is not clear which cell is infected first (15). Some studies have indicated that EBV first infects the epithelial cells of the pharynx via saliva, and later infects B lymphocytes after it has self-replicated; however, it has also been argued that B lymphocytes may succumb to infection before epithelial cells (16). Both situations can co-exist. The manner in which EBV infects these two target cells differs. EBV usually infects oral epithelial cells. The lymphocyte-rich tongue and tonsil tissues in the oral cavity present the EBV in oral epithelial cells with the opportunity to infect nearby B lymphocytes after replication (17). EBV infection of B lymphocytes does not directly lead to the development of HL; rather, the virus enters a stage of latent infection, during which the infected B lymphocytes undergo lytic replication to renew the infected B lymphocytes in the surrounding gaps. Meanwhile, the release of EBV in saliva can infect epithelial cells in a more efficient manner (18). Thus, by shuttling back and forth between B lymphocytes and epithelial cells (19), the life cycle of EBV is repeated (16). As a result, individuals with normal immunity are asymptomatic, while immunocompromised patients may develop EBV-related diseases (5,20). Möhl et al. found that EBV binds to its target cells in different ways: it infects B lymphocytes through endocytosis and infects epithelial cells through direct fusion with cellular membranes. Interestingly, no matter which cell is infected first or which infection pattern is applied, gp42 protein is always involved (10). Therefore, vaccines and treatments to fight EBV combine B-cell- and epithelial-cell-neutralizing antibodies to effectively block the entry of EBV via oral mucosa.
Compared with the pathogenesis of the influenza virus and HIV, the process of EBV infection of host cells is more complicated. During EBV infection, the membrane fusion and entry of the virus into cells are divided into multiple steps by the glycoproteins on its surface (21). Six EBV glycoproteins are typically found on the viral surface: gp350, gp220, gH, gL, gp42, and gB. Although the combinations of gp350 and gp220 with CR2 (CD21) can improve the efficiency of virus entry into host cells, they are not necessary for cell entry (22). The main function of gB, the most conserved glycoprotein, is to promote the maturity of EBV, and it is necessary for cell entry (23); however, it has been proposed that gB promotes virus-cell membrane fusion only under the regulation of gH/gL (24). The gp42 protein plays a central role in membrane fusion. As a directional switch (25), it can promote the fusion of EBV with B cells but inhibit the fusion of EBV with epithelial cells under the regulation of gH/gL proteins (23,26). However, the infected epithelial cells may be induced to express more HLA-II receptors and contain more gp42 (12). Importantly, the EBV-infected epithelial cells secrete some proteins (e.g., miR200s) that can stimulate the transformation of memory B lymphocytes from latent infection to proliferation, which is a basic step in the pathogenesis of HL (16).
The main function of HLA-II receptors is to present antigen (27). These receptors are predominantly expressed in conventional antigen-presenting cells including B cells, T cells, macrophages, and dendritic cells. Epithelial and endothelial cells can also be stimulated to express HLA-II receptors. The three different HLA-II molecules are HLA-DP, -DQ, and -DR (28). Each of these genotypes plays a critical role in EBV infection (29). In 2000, Haan et al. found the transient expression of HLA-DQ rendered EBV entry, and further research showed that only the HLA-DQβ*02 locus could regulate EBV entry (30). In 2002, Mullen et al. showed that the gp42 protein of EBV interacted with HLA-DR1 at a resolution of 2.65 Å (13). Li et al. found that EBV infection of B cells could be impaired by blocking the interaction between gp42 and HLA-II molecules or blocking the conversion of gp42 into sgp42 (29). There is a hypothesis that the conformation of gp42 changes slightly after HLA-II molecules bind to gp42, which rearranges the relevant glycoproteins in the viral envelope and alters the infection process, suggesting that gp42 plays a key role during EBV infection (31).
Our current research confirmed that the β-chain of HLA-DPB1 binds to the α-chain of gp42 to form a complex, the secondary structure of which is stabilized by hydrogen bonds. Figure 2D and Table 1 present the 3D structure diagram of the binding pattern and the contact areas between these two chains, respectively. In addition, the transfection efficiency of HLA-DPB1 in T1 cells (B lymphocytes) and 293T cells (epithelial cells) was measured using laboratory technology and the binding rate of 293T plasmid vector to gp42 was calculated.
Unfortunately, due to the low proportion of EGFP-expressing T1 cells during the construction of the HLA-DPB1 vectors, the binding rate of T1 cells to gp42 was not further detected. Limited by our experimental conditions, we were unable to obtain B lymphocytes that neither expressed HLA-DPB1 nor carried EBV genes. Consequently, our subsequent experiment only revealed that the binding efficiency of HLA-DPB1 to gp42 in 293T cells was 26–31.3%. In conclusion, the results of molecular dynamics simulation and laboratory tests in this study may help to improve our understanding of the pathogenic mechanism of HL caused by EBV infection of epithelial cells and B lymphocytes. Furthermore, tumorigenesis may be prevented by breaking hydrogen bonds, preventing hydrophilic binding, suppressing EBV activation, and lowering the concentration of gp42, and these strategies may also pave the way for effective therapies for treating HL.
Acknowledgments
Funding: Supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2070
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2070). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Correia S, Bridges R, Wegner F, et al. Sequence Variation of Epstein-Barr Virus: Viral Types, Geography, Codon Usage, and Diseases. J Virol 2018;92:e01132-18. [Crossref] [PubMed]
- Chen RW. Is there a place for the combination of brentuximab vedotin and bendamustine in treatment of patients with relapsed/refractory Hodgkin lymphoma? Ann Transl Med 2018;6:238. [Crossref] [PubMed]
- M A. Natural Killer cell transcriptome during primary EBV infection and EBV associated Hodgkin Lymphoma in children-A preliminary observation. Immunobiology 2020;225:151907. [Crossref] [PubMed]
- Liu ZL, Bi XW, Liu PP, et al. The Clinical Utility of Circulating Epstein-Barr Virus DNA Concentrations in NK/T-Cell Lymphoma: A Meta-Analysis. Dis Markers 2018;2018:1961058.
- Saha A, Robertson ES. Mechanisms of B-Cell Oncogenesis Induced by Epstein-Barr Virus. J Virol 2019;93:e00238-19. [Crossref] [PubMed]
- Chen YF, Chang CH, Huang ZN, et al. The JAK inhibitor antcin H exhibits direct anticancer activity while enhancing chemotherapy against LMP1-expressed lymphoma. Leuk Lymphoma 2019;60:1193-203. [Crossref] [PubMed]
- Ahmed W, Tariq S, Khan G. Tracking EBV-encoded RNAs (EBERs) from the nucleus to the excreted exosomes of B-lymphocytes. Sci Rep 2018;8:15438. [Crossref] [PubMed]
- Smatti MK, Al-Sadeq DW, Ali NH, et al. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front Oncol 2018;8:211. [Crossref] [PubMed]
- Bu W, Joyce MG, Nguyen H, et al. Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity 2019;50:1305-1316.e6. [Crossref] [PubMed]
- Möhl BS, Chen J, Longnecker R. Gammaherpesvirus entry and fusion: A tale how two human pathogenic viruses enter their host cells. Adv Virus Res 2019;104:313-43. [Crossref] [PubMed]
- Snijder J, Ortego MS, Weidle C, et al. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity 2018;48:799-811.e9. [Crossref] [PubMed]
- Trier N, Izarzugaza J, Chailyan A, et al. Human MHC-II with Shared Epitope Motifs Are Optimal Epstein-Barr Virus Glycoprotein 42 Ligands-Relation to Rheumatoid Arthritis. Int J Mol Sci 2018;19:317. [Crossref] [PubMed]
- Mullen MM, Haan KM, Longnecker R, et al. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell 2002;9:375-85. [Crossref] [PubMed]
- Morishima S, Shiina T, Suzuki S, et al. Evolutionary basis of HLA-DPB1 alleles affects acute GVHD in unrelated donor stem cell transplantation. Blood 2018;131:808-17. [Crossref] [PubMed]
- Tsao SW, Tsang CM, Lo KW. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci 2017;372:20160270. [Crossref] [PubMed]
- Lin Z, Swan K, Zhang X, et al. Secreted Oral Epithelial Cell Membrane Vesicles Induce Epstein-Barr Virus Reactivation in Latently Infected B Cells. J Virol 2016;90:3469-79. [Crossref] [PubMed]
- Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proc Natl Acad Sci U S A 2009;106:20464-9. [Crossref] [PubMed]
- Möhl BS, Chen J, Park SJ, et al. Epstein-Barr Virus Fusion with Epithelial Cells Triggered by gB Is Restricted by a gL Glycosylation Site. J Virol 2017;91:e01255-17. [Crossref] [PubMed]
- Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med 2002;8:594-9. [Crossref] [PubMed]
- Maurmann S, Fricke L, Wagner HJ, et al. Molecular parameters for precise diagnosis of asymptomatic Epstein-Barr virus reactivation in healthy carriers. J Clin Microbiol 2003;41:5419-28. [Crossref] [PubMed]
- Sathiyamoorthy K, Hu YX, Möhl BS, et al. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 2016;7:13557. [Crossref] [PubMed]
- Kanekiyo M, Bu W, Joyce MG, et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015;162:1090-100. [Crossref] [PubMed]
- Plate AE, Reimer JJ, Jardetzky TS, et al. Mapping regions of Epstein-Barr virus (EBV) glycoprotein B (gB) important for fusion function with gH/gL. Virology 2011;413:26-38. [Crossref] [PubMed]
- Chen J, Jardetzky TS, Longnecker R. The Cytoplasmic Tail Domain of Epstein-Barr Virus gH Regulates Membrane Fusion Activity through Altering gH Binding to gp42 and Epithelial Cell Attachment. mBio 2016;7:e01871-16. [Crossref] [PubMed]
- Haan KM, Lee SK, Longnecker R. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 2001;290:106-14. [Crossref] [PubMed]
- Sathiyamoorthy K, Jiang J, Möhl BS, et al. Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc Natl Acad Sci U S A 2017;114:E8703-E8710. [Crossref] [PubMed]
- Turrini R, Merlo A, Dolcetti R, et al. Differential down-modulation of HLA class I and II molecule expression on human tumor cell lines upon in vivo transfer. Cancer Immunol Immunother 2011;60:1639-45. [Crossref] [PubMed]
- Haan KM, Kwok WW, Longnecker R, et al. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J Virol 2000;74:2451-4. [Crossref] [PubMed]
- Li Q, Bu W, Gabriel E, et al. HLA-DQ β1 alleles associated with Epstein-Barr virus (EBV) infectivity and EBV gp42 binding to cells. JCI Insight 2017;2:e85687. [Crossref] [PubMed]
- Haan KM, Longnecker R. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc Natl Acad Sci U S A 2000;97:9252-7. [Crossref] [PubMed]
- Kirschner AN, Omerovic J, Popov B, et al. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol 2006;80:9444-54. [Crossref] [PubMed]