
Establishment of a prognostic model for predicting short-term disease-free survival in cases of hepatitis B-related hepatocellular carcinoma with the TP53 249Ser mutation in southern China
Introduction
Primary liver cancer is one of the most prevalent malignant tumors, with almost 841,000 new cases and 782,000 deaths occurring worldwide in 2018, and hepatocellular carcinoma (HCC) is the pathological type which accounts for approximately 75–85% of primary liver cancer (1). The major risk factors of HCC include: chronic infection with hepatitis virus (mainly hepatitis B and C), alcoholic/non-alcoholic liver disease, liver fluke infection, environmental carcinogens (such as aflatoxin) and genetic factors (2). According to a previously-published study, approximately 80% of cases of HCC arise in patients with chronic hepatitis B virus (HBV) infection in China and Africa (3).
The Guangxi region is located in southern China, where the main primary epidemiological factors of HCC are chronic HBV infection and aflatoxin exposure, which are recognized as leading carcinogenic factors in human HCC. As shown in previous molecular epidemiological investigations, the G-T mutation of TP53 249Ser is a recognized fingerprint of aflatoxin B1 (AFB1) exposure in genetic material of the local population (4-6). Previous studies have shown that the TP53 249Ser mutation is a high-frequency mutation hotspot in HCC patients from high-exposure areas of AFB1, such as Qidong, Guangxi, and South Africa (6-8).
The TP53 gene is known as a tumor suppressor, which plays an important role in cell growth and proliferation, cell cycle arrest, apoptosis, DNA repair, and senescence (9). TP53 gene mutation is the most common genetic mutation and is related to alteration of biological activity in cancer (10). Several studies have found that the TP53 gene mutation is also closely related to the clinical prognosis of HCC patients (11-13). However, there has been no research into the clinical prognostic value of TP53 249Ser in the HBV-related subtype of HCC in high-HBV infection and AFB1-exposure areas.
The nomogram is a commonly-used medical prediction model for predicting the likelihood of events such as DFS in an individual cancer patient (14,15). It has been reported that the nomogram achieved an optimal preoperative prediction of microvascular invasion (MVI) in HBV-related HCC (16). Therefore, in the present study, we mapped the TP53 249Ser mutation spectrum, and attempted to establish a prognostic model for predicting DFS in HCC cases in a high-HBV infection and high-aflatoxin exposure area. This is of great significance for improving the effect of therapy for HCC and predicting its clinical efficacy.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2788).
Methods
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University [2016(K-Y-E-011)] and informed consent was taken from all the patients.
Study population
A total of 485 patients with HCC who underwent hepatectomy at the Hepatobiliary Surgery Department of the First Affiliated Hospital of Guangxi Medical University (Nanning, China) between 2001 and 2013 were enrolled. Serum hepatitis B surface antigen was positive in all patients. The clinicopathological features of patients were identified from medical records and pathology reports, including age, gender, smoking status, drinking status, pathological grade features, preoperative serum alpha fetoprotein (AFP) levels, hepatic cirrhosis, radical resection, adjuvant antiviral therapy, and adjuvant transcatheter arterial chemoembolization (TACE). The Barcelona Clinic Liver Cancer (BCLC) staging system (17) was applied to the clinical stage of HCC. Child-Pugh class (18), portal vein tumor thrombosis (PVTT) (19) and radical resection (20) were as previously defined.
Follow-up
All patients’ follow-up information was obtained through outpatient interviews, telephone communication, and by reviewing medical records and hospital records. Follow-up continued until November 2015 unless terminated earlier due to death or recurrence events. Overall survival (OS) time was defined as from liver resection to HCC-related death. Disease-free survival (DFS) time was defined as from liver resection to HCC recurrence or distant metastasis. Patients still alive at the final follow-up were defined as censored.
DNA extraction and detection of TP53 249Ser mutation
All HCC specimens were collected within 1 hour of surgical resection and stored in an ultra-low temperature freezer at −80 °C until DNA extraction. DNA was extracted according to our previous method (21). TP53 249Ser mutations were detected by Sanger DNA sequencing after PCR amplification using the following primers: forward primer 5'-CTTGCCACAGGTCTCCCCAA-3'; reverse primer 5'-AGGGGTCAGAGGCAAGCAGA-3'. All PCR products were subjected to bidirectional sequencing using the ABI Prism 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA), by Shanghai Sangon Biological Engineering Technology & Services (Shanghai, China).
Immunohistochemistry and scoring
All paraffin-embedded HCC tissues were used for immunohistochemical staining of TP53 according to a previously-described method (22). The stained HCC sections were reviewed and scored by two pathologists independently who were blinded to clinical characteristics. At least ten fields were randomly selected at high-power (×400 magnification) at regions distant from necrotic areas, and the percentage of positive cells was calculated using the following formula: number of positive cells/total number of cells ×100%. Positive cells had brown granules in their nuclei. Scoring was performed according to previous criteria (23,24), and positive TP53 expression was defined as the presence of ≥10% positive cancer cells.
Propensity score matching (PSM)
To reduce the selectivity bias and heterogeneity between the two groups, we included the additional factor of a P value less than 0.1 in univariate analysis for PSM analysis (25). A 1:1 matching requirement by the nearest-neighbor matching algorithm without replacement was performed to select matched pairs of HBV-related HCC patients. SPSS 18.0 statistical software with R version 2.8.1 was employed to complete the PSM analysis.
Establishment of the nomogram
We performed PSM and Cox proportional hazards regression (CPHR) to identify independent risk factors for DFS. Next, a nomogram was constructed based on the results of multivariate logistic regression analysis, and formulated using the rms package of R, version 3.0 (http://www.r-project.org/). Each variable in the nomogram was based on scaling each regression coefficient in multiple logistic regression on a scale of 0 to 100 points. Furthermore, we used receiver operating characteristic curve analysis to calculate maximizing the Youden index (sensitivity + specificity −1) to determine the optimal cutoff point. Finally, the accuracy of the prediction model was estimated by specificity, sensitivity and likelihood ratio.
Statistical analysis
Univariate logistic regression analysis was used to compare the differences in clinical factors between the TP53 249Ser mutation group and the non-mutation group. The odds ratio (OR) and 95% confidence interval (95% CI) were used to evaluate the association between clinical factors and TP53 249Ser mutation. A Kaplan-Meier plot, univariate and multivariate Cox proportional hazards models were used to assess the association between clinical factors and clinical outcomes. The hazard ratio (HR) and the 95% CI were used to assess the correlation between clinical factors and clinical outcomes. All of the statistical analyses used in this study were performed using SPSS 18.0 (SPSS Inc., Chicago, IL, USA), and P<0.05 was considered statistically significant. GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA) was used to create statistical graphics.
Results
The spectrum of TP53 249Ser mutation in HBV-related HCC
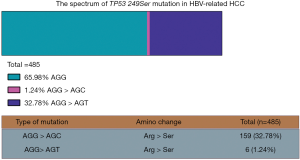
As shown in Figure 1, we identified 165 (34.02%) cases of TP53 249Ser mutation among the 485 HCC patients in this study. The main mutation types were AGG > AGT (Arg > Ser) (159 cases, 32.78%) and AGG > AGC (Arg > Ser) (six cases, 1.24%). These results are similar to our previously-published results (6).
Patient characteristics and PSM
A total of 485 HCC patients were enrolled, comprising 430 men and 55 women. The median age was 46 years. The patients were divided into the mutation group (n=165) and the non-mutation group (n=320) according to the TP53 249Ser mutation status (Table 1). In order to minimize the selection bias between the mutation group and the non-mutation group, PSM was estimated using univariate regression analysis including the covariates with P values less than 0.1 in Table 1. With a 1:1 ratio of propensity scoring, a total of 322 cases were included in the analysis, 161 cases from each group (Table 2). We found that median OS was 57 (non-mutation group) and 42 (mutation group) months. Median DFS was 11 (non-mutation group) and 6 months (mutation group). Our result showed that positive TP53 gene expression was significantly associated with TP53 249Ser mutation (P<0.001, Table 2)
Table 1
| Variable | TP53 249Ser mutation | |||
|---|---|---|---|---|
| Non-mutation group (n=320) | Mutation group (n=165) | OR* (95% CI) | P value* | |
| Age (years) | ||||
| ≤46 | 178 | 82 | 1 | |
| >46 | 142 | 83 | 1.17 (0.64–2.15) | 0.605 |
| Gender | ||||
| Male | 282 | 148 | 1 | |
| Female | 38 | 17 | 0.79 (0.54–1.15) | 0.215 |
| Race | ||||
| Han | 211 | 109 | 1 | |
| Minority | 109 | 56 | 0.72 (0.49–1.06) | 0.940 |
| BMI | ||||
| ≤25 | 265 | 136 | 1 | |
| >25 | 55 | 29 | 1.03 (0.63–1.69) | 0.915 |
| Smoking status | ||||
| None | 211 | 107 | 1 | |
| Ever | 109 | 58 | 1.05 (0.71–1.56) | 0.811 |
| Drinking status | ||||
| None | 198 | 97 | 1 | |
| Ever | 122 | 68 | 1.14 (0.78–1.67) | 0.509 |
| Child-Pugh class | ||||
| A | 267 | 136 | 1 | |
| B | 53 | 29 | 1.07 (0.65–1.77) | 0.778 |
| BCLC stage | ||||
| A | 189 | 95 | 1 | |
| B | 47 | 33 | 1.40 (0.84–2.32) | |
| C | 84 | 37 | 0.88 (0.55–1.39) | 0.238 |
| TP53 expressiona | ||||
| Negative | 133 | 26 | 1 | |
| Positive | 132 | 115 | 4.46 (2.73–7.27) | <0.001 |
| TACE status | ||||
| Before hepatectomy | ||||
| No | 244 | 138 | 1 | |
| Yes | 76 | 27 | 0.63 (0.39–1.02) | 0.061 |
| After hepatectomy | ||||
| No | 137 | 75 | 1 | |
| Yes | 183 | 90 | 0.90 (0.62–1.31) | 0.578 |
| Cirrhosis | ||||
| No | 37 | 22 | 1 | |
| Yes | 283 | 143 | 0.83 (0.47–1.46) | 0.511 |
| Serum AFPb | ||||
| ≤400 (ng/mL) | 165 | 82 | 1 | |
| >400 (ng/mL) | 134 | 68 | 1.02 (0.69–1.51) | 0.917 |
| Radical resectionc | ||||
| Yes | 166 | 102 | 1 | |
| No | 146 | 59 | 1.52 (1.03–2.25) | 0.035 |
| Pathological graded | ||||
| Good | 22 | 5 | 2.28 (0.84–6.17) | |
| Moderate | 245 | 127 | 1.96 (1.43–9.00) | 0.262 |
| Poor | 9 | 4 | ||
| Antiviral therapy | ||||
| No | 210 | 105 | 1 | |
| Yes | 110 | 60 | 1.09 (0.74–1.62) | 0.664 |
| Oncological behavior | ||||
| Tumor size | ||||
| ≤5 cm | 103 | 52 | 1 | |
| >5 cm | 217 | 113 | 1.03 (0.69–1.54) | 0.880 |
| No. of tumors | ||||
| Single (n=1) | 238 | 118 | 1 | |
| Multiple (n>1) | 82 | 47 | 1.16 (0.76–1.76) | 0.500 |
| Capsule | ||||
| Complete | 134 | 64 | 1 | |
| Incomplete | 129 | 76 | 0.92 (0.53–1.60) | |
| Absence | 57 | 25 | 1.23 (0.82–1.86) | 0.459 |
| Regional invasion | ||||
| Absence | 272 | 140 | 1 | |
| Presence | 48 | 25 | 1.01 (0.60–1.71) | 0.965 |
| Intrahepatic metastasis | ||||
| Absence | 146 | 75 | 1 | |
| Presence | 174 | 90 | 1.01 (0.69–1.47) | 0.972 |
| Vascular invasion | ||||
| Absence | 260 | 139 | 1 | |
| Presence | 60 | 26 | 0.81 (0.49–1.34) | 0.414 |
| PVTT | ||||
| No | 269 | 140 | 1 | |
| vp1 | 5 | 6 | 2.31 (0.69–7.69) | |
| vp2 | 14 | 3 | 0.41 (0.12–1.46) | |
| vp3 | 27 | 13 | 0.93 (0.46–1.85) | |
| vp4 | 5 | 3 | 1.15 (0.27–4.89) | 0.414 |
a, TP53 expression information was unavailable for 79 patients. b, AFP information was unavailable for 36 patients. c, radical resection information was unavailable for 12 patients. d, pathological grade information was unavailable for 73 patients. *, OR and P value for univariate analysis of logistic regression model. AFP, alpha-fetoprotein; TACE, transarterial chemoembolization; BMI, body mass index; PVTT, portal vein tumor thrombus; MST, median survival time; MRT, median recurrence time; HR, hazard ratio; 95% CI, 95% confidence interval.
Table 2
| Variable | TP53 249Ser mutation | |||
|---|---|---|---|---|
| Non-mutation group (n=161) | Mutation group (n=161) | OR* (95% CI) | P value* | |
| Age (years) | ||||
| ≤46 | 81 | 81 | 1 | |
| >46 | 80 | 80 | 1.00 (0.65–1.55) | 1.000 |
| Gender | ||||
| Male | 140 | 144 | 1 | |
| Female | 21 | 17 | 0.79 (0.40–1.55) | 0.490 |
| Race | ||||
| Han | 102 | 93 | 1 | |
| Minority | 59 | 68 | 1.26 (0.81–1.98) | 0.305 |
| BMI | ||||
| ≤25 | 132 | 133 | 1 | |
| >25 | 29 | 28 | 0.96 (0.54–1.70) | 0.884 |
| Smoking status | ||||
| None | 106 | 104 | 1 | |
| Ever | 55 | 57 | 1.06 (0.67–1.67) | 0.815 |
| Drinking status | ||||
| None | 102 | 93 | 1 | |
| Ever | 59 | 68 | 1.26 (0.81–1.98) | 0.305 |
| Child-Pugh class | ||||
| A | 135 | 126 | 1 | |
| B | 26 | 35 | 1.43 (0.74–2.76) | 0.288 |
| BCLC stage | ||||
| A | 105 | 95 | 1 | |
| B | 21 | 29 | 1.53 (0.82–2.85) | |
| C | 35 | 37 | 1.17 (0.68–2.00) | 0.402 |
| TP53 expressiona | ||||
| Negative | 69 | 25 | 1 | |
| Positive | 67 | 112 | 4.61 (2.67–8.00) | <0.001 |
| TACE status | ||||
| Before hepatectomy | ||||
| No | 122 | 135 | 1 | |
| Yes | 39 | 26 | 0.60 (0.35–1.05) | 0.073 |
| After hepatectomy | ||||
| No | 71 | 73 | 1 | |
| Yes | 90 | 88 | 0.95 (0.61–1.48) | 0.823 |
| Cirrhosis | ||||
| No | 15 | 22 | 1 | |
| Yes | 146 | 139 | 0.65 (0.324–1.30) | 0.224 |
| Serum AFPb | ||||
| ≤400 (ng/mL) | 85 | 80 | 1 | |
| >400 (ng/mL) | 66 | 66 | 1.06 (0.67–1.68) | 0.795 |
| Radical resection | ||||
| Yes | 102 | 102 | 1 | |
| No | 59 | 59 | 1.00 (0.64–1.57) | 1.000 |
| Pathological gradec | ||||
| Good | 14 | 5 | 1 | |
| Moderate | 127 | 124 | 2.73 (0.96–7.82) | |
| Poor | 7 | 4 | 1.60 (0.34–7.90) | 0.130 |
| Antiviral therapy | ||||
| No | 111 | 103 | 1 | |
| Yes | 50 | 58 | 1.30 (0.82–2.06) | 0.273 |
| Oncological behavior | ||||
| Tumor size | ||||
| ≤5 cm | 56 | 50 | 1 | |
| >5 cm | 105 | 111 | 1.18 (0.74–1.88) | 0.477 |
| No. of tumors | ||||
| Single (n=1) | 122 | 118 | 1 | |
| Multiple (n>1) | 39 | 43 | 1.14 (0.69–1.88) | 0.609 |
| Capsule | ||||
| Complete | 63 | 61 | 1 | |
| Incomplete | 72 | 75 | 0.99 (0.52–1.91) | |
| Absence | 26 | 25 | 1.08 (0.67–1.74) | 0.945 |
| Regional invasion | ||||
| Absence | 139 | 136 | 1 | |
| Presence | 22 | 25 | 1.16 (0.63–2.16) | 0.636 |
| Intrahepatic metastasis | ||||
| Absence | 70 | 73 | 1 | |
| Presence | 91 | 88 | 0.93 (0.60–1.44) | 0.737 |
| Vascular invasion | ||||
| Absence | 135 | 135 | 1 | |
| Presence | 26 | 26 | 1.00 (0.55–1.81) | 1.000 |
| PVTT | ||||
| No | 140 | 136 | 1 | |
| vp1 | 0 | 6 | NA | |
| vp2 | 8 | 3 | 0.39 (0.10–1.49) | |
| vp3 | 11 | 13 | 1.22 (0.53–2.81) | |
| vp4 | 2 | 3 | 1.54 (0.25–9.39) | 0.657 |
a, TP53 expression information was unavailable for 49 patients. b, AFP information was unavailable for 25 patients. c, pathological grade information was unavailable for 41 patients. *, OR and P value for univariate analysis of logistic regression model. AFP, alpha-fetoprotein; TACE, transarterial chemoembolization; BMI, body mass index; PVTT, portal vein tumor thrombus; OR, odds ratio; 95% CI, 95% confidence interval.
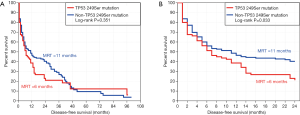
The results of Kaplan-Meier survival analysis (Figure 2) and univariate CPHR analysis only showed a statistically-significant difference in 2-year DFS between the two groups (P=0.033, HR =1.40, 95% CI: 1.01–1.94), but there was no significant difference in long-term DFS (P=0.351, HR =1.16, 95% CI: 0.83–1.63).
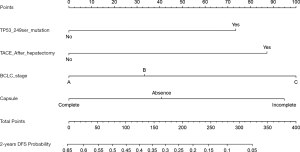
Prognostic model for 2-year DFS
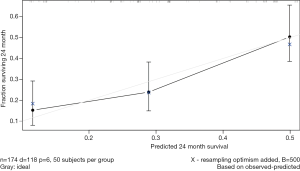
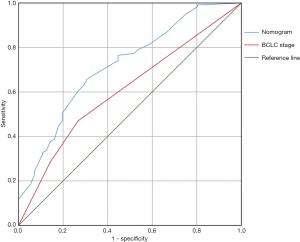
The results of univariate CPHR analysis are shown in Table 3. All significant indicators were then incorporated into multivariate CPHR. As shown in Table 4, we found that TACE after surgery, status of TP53 249Ser mutation, BCLC staging and tumor capsule were independent prognostic factors for 2-year DFS (P<0.05). We constructed the independent predictors above into a nomogram (Figure 3), and then used the bootstrap validation method to internally validate the resulting model (26) (Figure 4). The results indicated that the nomogram showed a good accuracy in assessing 2-year DFS, with a C-index of 0.718 (95% CI: 0.638–0.799), which was significantly greater than that of the BCLC staging system (C-index: 0.606, 95% CI: 0.519–0.693) (Figure 5). The optimal cutoff point of the total nomogram scores was determined to be 160, and the specificity and sensitivity were 65.5% and 69.6% respectively.
Table 3
| Variables | Patients (n=322) | Overall survival | Disease-free survival | |||||
|---|---|---|---|---|---|---|---|---|
| MST (months) | HR* (95% CI) | P* | MRT (months) | HR* (95% CI) | P* | |||
| TP53 249Ser mutation | ||||||||
| Non-mutation | 161 | 57 | 1 | 11 | 1 | |||
| Mutation | 161 | 42 | 1.27 (0.92–1.75) | 0.146 | 6 | 1.16 (0.83–1.63) | 0.351 | |
| Age (years) | ||||||||
| ≤46 | 162 | 57 | 1 | 7 | 1 | |||
| >46 | 160 | 48 | 0.88 (0.64–1.21) | 0.438 | 12 | 0.78 (0.56–1.09) | 0.151 | |
| Gender | ||||||||
| Male | 284 | 48 | 1 | 7 | 1 | |||
| Female | 38 | 51 | 0.66 (0.366–1.19) | 0.166 | 12 | 0.89 (0.52–1.53) | 0.673 | |
| Race | ||||||||
| Han | 195 | 48 | 1 | 8 | 1 | |||
| Minority | 127 | 51 | 1.03 (0.74–1.43) | 0.868 | 8 | 1.22 (0.87–1.72) | 0.243 | |
| BMI | ||||||||
| ≤25 | 265 | 45 | 1 | 9 | 1 | |||
| >25 | 57 | 51 | 0.93 (0.61–1.40) | 0.719 | 7 | 1.29 (0.88–1.89) | 0.195 | |
| Smoking status | ||||||||
| None | 210 | 61 | 1 | 9 | 1 | |||
| Ever | 112 | 40 | 1.20 (0.86–1.67) | 0.275 | 6 | 1.04 (0.73–1.47) | 0.839 | |
| Drinking status | ||||||||
| None | 195 | 57 | 1 | 8 | 1 | |||
| Ever | 127 | 41 | 1.26 (0.91–1.74) | 0.158 | 7 | 1.03 (0.73–1.45) | 0.873 | |
| TP53 expressiona | ||||||||
| Negative | 94 | 45 | 1 | 12 | 1 | |||
| Positive | 179 | 42 | 1.08 (0.75–1.58) | 0.673 | 6 | 1.04 (0.71–1.52) | 0.838 | |
| TACE status | ||||||||
| Before hepatectomy | ||||||||
| No | 257 | 57 | 1 | 9 | 1 | |||
| Yes | 65 | 44 | 1.07 (0.73–1.58) | 0.720 | 6 | 1.37 (0.88–1.95) | 0.190 | |
| After hepatectomy | ||||||||
| No | 144 | 88 | 1 | 14 | 1 | |||
| Yes | 178 | 42 | 1.16 (0.84–1.62) | 0.368 | 6 | 1.64 (1.13–2.40) | 0.010 | |
| BCLC stage | ||||||||
| A | 200 | 84 | 1 | <0.001 | 12 | 1 | 0.030 | |
| B | 50 | 71 | 1.63 (1.05–2.53) | 0.031 | 6 | 1.19 (0.75–1.90) | 0.456 | |
| C | 72 | 17 | 3.31 (2.29–4.78) | <0.001 | 2 | 1.70 (1.15–2.53) | 0.008 | |
| Child–Pugh class | ||||||||
| A | 261 | 57 | 1 | 7 | 1 | |||
| B | 61 | 34 | 1.57 (1.02–2.43) | 0.040 | 6 | 1.22 (0.75–2.00) | 0.428 | |
| Cirrhosis | ||||||||
| No | 37 | 88 | 1 | 38 | 1. | |||
| Yes | 285 | 45 | 1.55 (0.89–2.68) | 0.122 | 7 | 1.79 (0.94–3.41) | 0.077 | |
| Antiviral therapy | ||||||||
| No | 214 | 58 | 1 | 6 | 1 | |||
| Yes | 108 | NA | 0.54 (0.36–0.81) | 0.003 | 13 | 0.62 (0.44–0.90) | 0.010 | |
| AFP (ng/mL)b | ||||||||
| ≤400 | 165 | 61 | 1 | 12 | 1 | |||
| >400 | 132 | 41 | 1.22 (0.87–1.72) | 0.246 | 7 | 1.08 (0.76–1.54) | 0.661 | |
| Radical resection | ||||||||
| Yes | 204 | 71 | 1 | 11 | 1 | |||
| No | 118 | 36 | 1.59 (1.15–2.19) | 0.005 | 5 | 1.60 (1.14–2.24) | 0.007 | |
| Pathological gradec | ||||||||
| Good | 19 | NA | 1 | 0.496 | 6 | 1 | 0.164 | |
| Moderate | 251 | 48 | 1.61 (0.71–3.66) | 0.255 | 7 | 1.54 (0.67–3.56) | 0.308 | |
| Poor | 11 | NA | 1.33 (0.38–4.73) | 0.656 | 3 | 3.18 (0.95–10.68) | 0.061 | |
| Oncological features | ||||||||
| Tumor size | ||||||||
| ≤5 cm | 106 | 123 | 1 | 17 | 1 | |||
| >5 cm | 216 | 41 | 1.9 (1.31–2.82) | 0.001 | 5 | 1.81 (1.25–2.62) | 0.002 | |
| No. of tumors | ||||||||
| Single (n=1) | 240 | 51 | 1 | 11 | 1 | |||
| Multiple (n>1) | 82 | 39 | 1.38 (0.97–1.96) | 0.072 | 6 | 1.15 (0.79–1.68) | 0.456 | |
| Capsule | ||||||||
| Complete | 124 | 95 | 1 | 0.002 | 14 | 1 | 0.014 | |
| Incomplete | 147 | 36 | 1.51 (0.93–2.46) | 0.099 | 5 | 1.21 (0.71–2.06) | 0.489 | |
| Absence | 51 | 51 | 1.93 (1.33–2.78) | <0.001 | 8 | 1.72 (1.19–2.48) | 0.004 | |
| Regional invasion | ||||||||
| Absence | 275 | 57 | 1 | 11 | 1 | |||
| Presence | 47 | 40 | 1.32 (0.84–2.08) | 0.235 | 2 | 1.80 (1.17–2.77) | 0.008 | |
| Intrahepatic metastasis | ||||||||
| Absence | 179 | 75 | 1 | 12 | 1 | |||
| Presence | 143 | 36 | 1.60 (1.16–2.20) | 0.004 | 4 | 1.54 (1.10–2.15) | 0.012 | |
| Vascular invasion | ||||||||
| Absence | 270 | 73 | Ref. | 9 | 1 | |||
| Presence | 52 | 12 | 3.40 (2.32–5.00 | <0.001 | 2 | 1.57 (1.03–2.40) | 0.036 | |
| PVTT | ||||||||
| No | 276 | 71 | Ref. | <0.001 | 11 | 1 | 0.014 | |
| vp1 | 6 | 7 | 4.67 (1.70–12.84) | 0.003 | 1 | 2.29 (0.73–7.24) | 0.158 | |
| vp2 | 11 | 17 | 4.16 (1.53–6.52) | <0.002 | 2 | 2.24 (0.82–6.13) | 0.117 | |
| vp3 | 24 | 12 | 3.00 (1.80–5.00) | <0.001 | 3 | 1.43 (0.80–2.55) | 0.226 | |
| vp4 | 5 | 8 | 6.52 (2.37–17.94) | <0.001 | 1 | 4.49 (1.61–12.55) | 0.004 | |
P<0.05 is statistically significant. a, TP53 expression information was unavailable for 49 patients. b, AFP information was unavailable for 25 patients. c, pathological grade information was unavailable for 41 patients. *, HR and P value for univariate survival analysis of Cox proportional hazard regression model. AFP, alpha-fetoprotein; TACE, transarterial chemoembolization; BMI, body mass index; PVTT, portal vein tumor thrombus; MST, median survival time; MRT, median recurrence time; HR, hazard ratio; 95% CI, 95% confidence interval.
Table 4
| Variables | Patients (n=322) | 2 years disease free survival | ||
|---|---|---|---|---|
| MRT (months) | HR* (95% CI) | P* | ||
| TP53 249Ser mutation | ||||
| Non-mutation | 161 | 11 | 1 | |
| Mutation | 161 | 6 | 1.47 (1.02–2.18) | 0.039 |
| TACE after hepatectomy | ||||
| No | 144 | 9 | 1 | |
| Yes | 178 | 6 | 1.65 (1.08–2.52) | 0.020 |
| Capsule | ||||
| Complete | 124 | 14 | 1 | 0.044 |
| Incomplete | 147 | 5 | 1.27 (0.70–2.31) | 0.438 |
| Absence | 51 | 8 | 1.71 (1.12–2.61) | 0.013 |
| BCLC stage | ||||
| A | 200 | 12 | 1 | 0.012 |
| B | 50 | 6 | 1.35 (0.82–2.23) | 0.234 |
| C | 72 | 2 | 1.92 (1.25–2.95) | 0.003 |
P<0.05 is statistically significant. *, HR and P value for multivariate survival analysis of Cox proportional hazard regression model. MRT, median recurrence time; HR, hazard ratio; 95% CI, 95% confidence interval.
Conclusions
HCC is one of the most common malignant tumors in the world, and more than half of new cases occur in China every year (1). HBV infection and AFB1 exposure are two major risk factors of HCC occurrence (2). The Guangxi area, located in southern China, has a higher incidence of HCC than the national average due to its unique environmental background of high exposure to AFB1 and high levels of endemic HBV, both of which are currently recognized risk factors for human HCC. In addition, HBV infection and AFB1 exposure are known to have synergistic carcinogenic effects (27).
Studies on HCC patients with a background of AFB1 exposure found that tumors exhibited a high frequency of a G-T mutation at codon 249 of the TP53 gene, which is considered as the molecular fingerprint of AFB1 exposure during HCC pathogenesis in this region, revealing the interaction between environment and genes during hepatocarcinogenesis (6,28,29). Our previous studies also provided evidence that there was a hotspot for this characteristic TP53 mutation in HCC in Guangxi, in which the TP53 249Ser mutation rate in recurrent HCC was as high as 60% (30). Holmes et al. reported that the urine metabolism spectrum of a southern population, represented by Guangxi, was significantly different from that of northern China, indicating that diet and other environmental factors differed greatly between the northern and southern populations and that this in turn made a huge difference to metabolism (31). Previous study has shown that AFB1 contamination in food is an important factor underlying the high incidence of HCC in Guangxi (32). Previous reports also revealed that the exposure level gradient of AFB1 paralleled the incidence of HCC (33-35) and exposure level of aflatoxin is closely related to TP53 249Ser mutation and the occurrence of HCC (36,37).
In our study, positive TP53 gene expression was significantly associated with TP53 249Ser mutation (P<0.001, Table 2), and TP53 249Ser mutation was still significantly correlated with 2-year DFS of patients undergoing hepatectomy after correction of the clinical factors significantly associated with recurrence (P=0.039, HR =1.47, 95% CI: 1.02–2.18). This result indicates that patients with the TP53 249Ser mutation will have a higher risk of tumor recurrence after surgery compared with patients without the mutation. Therefore, TP53 249Ser mutation may be an independent risk factor for tumor recurrence within 2 years after hepatectomy in patients with HBV-related HCC. Currently, several studies on Japanese HCC patients have shown that mutations in the exon region of the TP53 gene are significantly correlated with short-term recurrence and pathological differentiation of tumors, a finding that is similar to the results of this study (37,38). The difference is that our localization is more specific, only targeting the codon 249 mutation of the TP53 gene. A study of HCC patients in Guangdong, a region neighboring Guangxi, demonstrated that TP53 gene 249Ser and V157F mutation hotspots were significantly correlated with short-term OS of HCC patients after surgery (11). Similarly, another report found that TP53 gene mutation indicated poor prognosis among HCC patients in Taiwan (39).
As a medical tool to predict clinical events, in the field of HCC, the nomogram has been used in many studies to establish models to predict the clinical prognosis of patients with liver cancer (40,41). In the present study, through the clinical pathological characteristics of HCC patients and based on univariate and multivariate CPHR analysis, we preliminarily established a nomogram to predict the short-term DFS of HCC patients with AFB1 and HBV exposure. In this nomogram, TACE after surgery, status of TP53 249Ser mutation, BCLC staging and tumor capsule were independent prognostic factors for 2-year DFS. In order to make the model applicable to the clinic, we determined the cutoff value of 160 to evaluate prognosis by calculating the maximum Youden index, but the sensitivity and specificity were not satisfactory. Therefore a score of more than 160 points was considered a high-risk group for tumor recurrence within two years, which allows the nomogram to serve as a tool for making rough predictions about the prognosis of patients after hepatectomy. Further evaluation revealed that the C-index of the nomogram (C-index: 0.718, 95% CI: 0.638–0.799) exceeded that of the BCLC staging system (P=0.024) (C-index: 0.606, 95% CI: 0.519–0.693). The reason may be that the liver disease background and environmental background of HCC patients in the region of this study were different from those in Europe. In addition the risk factors incorporated in the prediction model may be more geographically representative.
There were still some limitations in our research that need further improvement. The cases included in the study were from a single center and may not be sufficiently representative. The nomogram we established was based on the patient’s clinical characteristics. Furthermore, in vitro and in vivo studies are necessary to elucidate the molecular mechanism by which TP53 249Ser mutation affects the clinical prognosis of HBV-related HCC patients.
In this study, we found that the TP53 249Ser mutation may be a high risk factor of HBV-related HCC recurrence in the short term. Then we established a preliminary prediction model for 2-year DFS, based on regional characteristics, high AFB1 exposure and high incidence of chronic hepatitis B. This study not only fills a gap in the relevant research field but will also help with the implementation of active prevention and treatment strategies for high-risk populations in areas with high incidence of liver cancer.
Acknowledgments
The authors thank Prof. Cheryl A. Winkler and Prof. Stephen J. O’Brien for their invaluable help with our study. Thanks also go to Prof. Yanling Hu and Prof. Zengnan Mo from the Medical Scientific Research Center of Guangxi Medical University for their contribution to data analysis, and to the researchers Jiaquan Li and Ying Gui from Guangxi Medical University for their contribution to specimen management. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This work was supported in part by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2788
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2788
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2788). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University [2016(K-Y-E-011)] and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Fujiwara N, Friedman SL, Goossens N, et al. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018;68:526-49. [Crossref] [PubMed]
- Shen HM, Ong CN. Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat Res 1996;366:23-44. [Crossref] [PubMed]
- Liu ZM, Li LQ, Peng MH, et al. Hepatitis B virus infection contributes to oxidative stress in a population exposed to aflatoxin B1 and high-risk for hepatocellular carcinoma. Cancer Lett 2008;263:212-22. [Crossref] [PubMed]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Qi LN, Bai T, Chen ZS, et al. The p53 mutation spectrum in hepatocellular carcinoma from Guangxi, China: role of chronic hepatitis B virus infection and aflatoxin B1 exposure. Liver Int 2015;35:999-1009. [Crossref] [PubMed]
- Egal S, Hounsa A, Gong Y, et al. Dietary exposure to aflatoxin from maize and groundnut in young children from Benin and Togo, West Africa. Int J Food Microbiol 2005;104:215-24. [Crossref] [PubMed]
- Stern MC, Umbach DM, Mimi CY, et al. Hepatitis B, aflatoxin B1, and p53 codon 249 mutation in hepatocellular carcinomas from Guangxi, People’s Republic of China, and a meta-analysis of existing studies. Cancer Epidemiol Biomarkers Prev 2001;10:617-25. [PubMed]
- Hafner A, Bulyk ML, Jambhekar A, et al. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol 2019;20:199-210. [Crossref] [PubMed]
- Kim MP, Zhang Y, Lozano G. Mutant p53: Multiple Mechanisms Define Biologic Activity in Cancer. Front Oncol 2015;5:249. [Crossref] [PubMed]
- Su H, Zhao J, Xiong Y, et al. Large-scale analysis of the genetic and epigenetic alterations in hepatocellular carcinoma from Southeast China. Mutat Res 2008;641:27-35. [Crossref] [PubMed]
- Chaudhary K, Poirion OB, Lu L, et al. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res 2018;24:1248-59. [Crossref] [PubMed]
- Long J, Wang A, Bai Y, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 2019;42:363-74. [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-e180. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Lei Z, Li J, Wu D, et al. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus–related hepatocellular carcinoma within the milan criteria. JAMA Surg 2016;151:356-63. [Crossref] [PubMed]
- Forner A, Reig ME, de Lope CR, et al. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis 2010;30:61-74. [Crossref] [PubMed]
- Garcia-Tsao G, Abraldes JG, Berzigotti A, et al. Portal hypertensive bleeding in cirrhosis: Risk stratification, diagnosis, and management: 2016 practice guidance by the American Association for the study of liver diseases. Hepatology 2017;65:310-35. [Crossref] [PubMed]
- Kondo K, Chijiiwa K, Kai M, et al. Surgical strategy for hepatocellular carcinoma patients with portal vein tumor thrombus based on prognostic factors. J Gastrointest Surg 2009;13:1078-83. [Crossref] [PubMed]
- Yu L, Liu X, Han C, et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis B virus-related hepatocellular carcinoma after hepatectomy: a cohort study for 10 years’ follow up. Hepatol Res 2016;46:765-74. [Crossref] [PubMed]
- Han C, Yu L, Liu X, et al. ATXN7 gene variants and expression predict post-operative clinical outcomes in hepatitis B virus-related hepatocellular carcinoma. Cell Physiol Biochem 2016;39:2427-38. [Crossref] [PubMed]
- Han C, Liao X, Qin W, et al. EGFR and SYNE2 are associated with p21 expression and SYNE2 variants predict post-operative clinical outcomes in HBV-related hepatocellular carcinoma. Sci Rep 2016;6:31237. [Crossref] [PubMed]
- Cote RJ, Dunn MD, Chatterjee SJ, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res 1998;58:1090-4. [PubMed]
- Esrig D, Elmajian D, Groshen S, et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. N Engl J Med 1994;331:1259-64. [Crossref] [PubMed]
- Kuss O, Blettner M, Borgermann J. Propensity Score: an Alternative Method of Analyzing Treatment Effects. Dtsch Arztebl Int 2016;113:597-603. [PubMed]
- Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J 2014;35:1925-31. [Crossref] [PubMed]
- McCullough AK, Lloyd RS. Mechanisms underlying aflatoxin-associated mutagenesis - Implications in carcinogenesis. DNA Repair (Amst) 2019;77:76-86. [Crossref] [PubMed]
- Bressac B, Kew M, Wands J, et al. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 1991;350:429-31. [Crossref] [PubMed]
- Hsu IC, Metcalf R, Sun T, et al. Mutational hot spot in the p53 gene in human hepatocellular carcinomas. Nature 1991;350:427-8. [Crossref] [PubMed]
- Qi LN, Li LQ, Chen YY, et al. Genome-wide and differential proteomic analysis of hepatitis B virus and aflatoxin B1 related hepatocellular carcinoma in Guangxi, China. PloS One 2013;8:e83465. [Crossref] [PubMed]
- Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008;453:396-400. [Crossref] [PubMed]
- Yeh FS, Mimi CY, Mo CC, et al. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res 1989;49:2506-9. [PubMed]
- Tao P, Zhi-Ming L, Tang-Wei L, et al. Associated Factors in Modulating Aflatoxin B 1–Albumin Adduct Level in Three Chinese Populations. Dig Dis Sci 2005;50:525-32. [Crossref] [PubMed]
- Peng T, Li LQ, Peng MH, et al. Is correction for protein concentration appropriate for protein adduct dosimetry? Hypothesis and clues from an aflatoxin B1-exposed population. Cancer Sci 2007;98:140-6. [Crossref] [PubMed]
- Chu YJ, Yang HI, Wu HC, et al. Aflatoxin B1 exposure increases the risk of hepatocellular carcinoma associated with hepatitis C virus infection or alcohol consumption. Eur J Cancer 2018;94:37-46. [Crossref] [PubMed]
- Ozturk M. p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet 1991;338:1356-9. [Crossref] [PubMed]
- Nogueira JA, Ono-Nita SK, Nita ME, et al. 249 TP53 mutation has high prevalence and is correlated with larger and poorly differentiated HCC in Brazilian patients. BMC Cancer 2009;9:204. [Crossref] [PubMed]
- Hsu CY, Liu PH, Hsia CY, et al. Nomogram of the Barcelona Clinic Liver Cancer system for individual prognostic prediction in hepatocellular carcinoma. Liver Int 2016;36:1498-506. [Crossref] [PubMed]
- Yuan RH, Chang KT, Chen YL, et al. S100P expression is a novel prognostic factor in hepatocellular carcinoma and predicts survival in patients with high tumor stage or early recurrent tumors. PLoS One 2013;8:e65501. [Crossref] [PubMed]
- Ma KW, She WH, Cheung TT, et al. Validated nomogram for the prediction of disease-free survival after hepatectomy for hepatocellular carcinoma within the Milan criteria: individualizing a surveillance strategy. Surg Today 2019;49:521-8. [Crossref] [PubMed]
- Torzilli G, Donadon M, Belghiti J, et al. Predicting individual survival after hepatectomy for hepatocellular carcinoma: a novel nomogram from the “HCC East & West Study Group”. J Gastrointest Surg 2016;20:1154-62. [Crossref] [PubMed]


