
Risk assessment model and nomogram established by differentially expressed lncRNAs for early-stage lung squamous cell carcinoma
Introduction
Lung cancer is one of the most common malignancies that lead to high rates of morbidity and mortality. In 2018, 2,093,876 newly diagnosed cases of lung cancer were reported, being responsible for an estimated 1,761,007 deaths in the world (1). Additionally, patients with non-small cell lung cancer (NSCLC) account for approximately 85% of all lung cancer patients (2). Although therapeutic strategies (e.g., targeted therapies and immunotherapy) for treating advanced-stage NSCLC have made great progress over the last 10 years, no major breakthroughs have been achieved in the treatment of early-stage NSCLC in recent years (3). Furthermore, early-stage NSCLC can be surgical resection, but the 5-year survival rate of NSCLC was only 36–73% (4). Also, a large number of stage I and II patients still face the risk of recurrence after complete surgical resection. As a common subtype of NSCLC, lung SCC accounts for about 40% of all lung cancer and is associated with poor prognosis compared to lung adenocarcinoma (5,6). Moreover, a study has shown the enormous heterogeneity of genomic and epigenomic landscape between lung adenocarcinoma and lung SCC (7), targeted therapies for lung adenocarcinoma are largely ineffective against lung SCC (8). Besides, carcinoma cells of lung SCC often penetrate into the adjacent tissue easily, leading to a high reoccurrence of lung SCC, which makes the prognosis more difficult to assess (9). Accordingly, it is essential to develop new strategies in order to improve the prognosis of lung SCC.
In recent years, long noncoding RNAs (lncRNAs) have received much more attention. lncRNAs are a group of RNAs that are >200 nucleotides in length, with no protein-coding capacity (10). Many studies have demonstrated that lncRNAs are involved in the development and metastasis of a variety of cancers (11-13), including hepatocellular carcinoma (14), lung cancer (15), and colon cancer (16), etc. Also, some lncRNAs have been reported to be associated with the prognosis of lung cancer. For example, a previous study has identified a prognostic lncRNA (LL22NC03-N64E9.1) which was highly expressed in lung cancer tissues and related to improved survival time (17). Furthermore, several lncRNA signatures for predicting the prognosis of NSCLC or lung SCC have been established (9,10,18-21). However, no previous studies have investigated the association between lncRNAs-based risk assessment model and the prognosis of early-stage (stage I and II) lung SCC.
The purpose of this paper is to screen the differentially expressed lncRNAs of patients with stage I–II lung SCC in order to identify lncRNAs that could serve as markers for the prognosis of lung SCC. Moreover, we constructed two lncRNA-based prognostic models for predicting the survival of patients with stage I–II lung SCC.
Methods
Data and data processing
The raw data of lncRNA expression and corresponding clinical information of early-stage (stage I–II) lung SCC patients were obtained from The Cancer Genome Atlas (TCGA) websites (https://cancergenome.nih.gov/). According to samples’ ID, lncRNA expression data were then matched to available clinical data, including survival status, overall survival (OS) time, age, gender, TNM stage, T stage. Furthermore, samples without sufficient clinical information were excluded.
Screening differentially expressed lncRNAs
Differential expression analysis was performed using edgeR 3.10 package (www.bioconductor.org/packages/release/bioc/html/edgeR.html) in R software (version 3.6.0, www.r-project.org), and lncRNAs with a false discovery rate (FDR) <0.05 and a |log2 fold change (log2FC)| >2 were treated as differentially expressed lncRNAs (DELs). Subsequently, univariate Cox regression analysis was first performed to screen potential prognostic factors (lncRNAs).
Establishment of an lncRNAs-based risk assessment model
Those lncRNAs with a P value less than 0.05 in univariable Cox regression were further analyzed using least absolute shrinkage and selection operator (LASSO) model to obtain key lncRNAs. LASSO is a widely used approach for the regression of high-dimensional indicators. By using LASSO method, regression coefficients may be reduced to zero for some predictor variables (lncRNAs), which will be removed from the LASSO model. After that, most potential predictor variables were shrunk to zero, and only a small number of lncRNAs were left for further analysis. The key lncRNAs identified by the LASSO model were subjected to a multivariate Cox regression analysis.
Based on the results of multivariate Cox analysis, lncRNAs with statistical significance were further used to establish the risk assessment model, and the risk scores were calculated according to the formula produced by multivariate Cox regression model. Then, patients were divided into a high-risk level group and low-risk level group based on the median cutoff point of the risk scores. The “glmnet” package in R software 3.6.0 was utilized to perform the LASSO analysis. The results of multivariable Cox regression analysis were visualized by forest plot using “survminer” package.
Evaluation for the risk assessment model constructed by DELs
OS times among high-risk and low-risk groups were compared by the Kaplan-Meier (KM) method. To evaluate the performance of the lncRNA risk assessment model for survival prediction, time-dependent receiver-operating characteristic (ROC) curves were constructed and the area under the ROC curves (AUC) values were calculated. The ROC curves of 3‐ and 5‐year OS was performed using “timeROC” package of R software.
Construction and assessment of a risk level-based nomogram
Before the construction of the risk level-based nomogram, clinical variables with missing values were excluded. Subsequently, the possible prognostic factors, including age, gender, TNM stage, T stage, and risk level were submitted for univariate Cox regression analysis, respectively. Besides, optimized threshold values for age were determined using the X-Tile software (Version 3.6.1, Yale University). Clinical factors with a P value <0.5 were further included in multivariate Cox regression model.
The nomogram based on the multivariate Cox regression model was formulated to predict 3‐ and 5‐year OS of stage I–II lung SCC. Calibration plots were utilized to assess the performance of the nomogram. A calibration plot reports predicted probabilities of survival (on the x-axis) against observed outcome frequencies (on the y-axis), and 45-degree line represents the perfect prediction. Also, predictive accuracy of the nomogram was measured by AUC of the ROC curves. The nomogram and calibration plots were built using R package “rms”, and ROC curves was performed using “timeROC” package of R software.
KM Survival analysis
KM method (log-rank test) was applied to analyze the OS time by various factors, including age, T stage, and potential prognostic lncRNAs identified by multivariate Cox regression analysis.
Statistical analysis
In the present study, primary end points were OS defined as death from any cause, and hazard ratios (HR) with 95% confidence intervals (95% CI) of the prognostic factors were computed based on the Cox regression model. A P value <0.05 was considered to indicate a statistically significant difference. Additionally, the AUC was used to evaluate the predictive capacity and accuracy of the risk assessment model and nomogram. An AUC of 0.51–0.6 is considered poor performance, 0.61–0.7 was general, 0.71–0.9 was moderate, and >0.9 was excellent. All analysis was carried out through using R software (version 3.6.0), and Cox regression analysis as well as the KM survival analysis was performed using “survival” package of R software.
Results
Differentially expression lncRNAs
After excluding 11 patients without age, survival data or enough clinical information, the lncRNAs data of 395 patients with stage I–II lung SCC were finally retained for further analysis. The lncRNAs expression levels were compared between 49 normal and 395 tumor tissues (stage I–II lung SCC), and then a total of 2,021 DELs were obtained. Significant lncRNA expression changes were visualized using a volcano plot (Figure 1A).
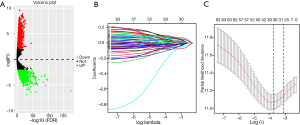
A 5-lncRNA-based risk assessment model
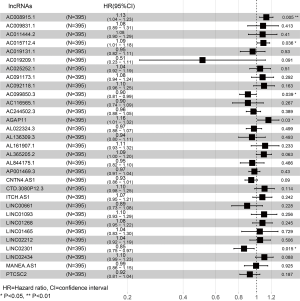
Among these DELs, 63 potential prognostic lncRNAs were identified by univariate Cox regression analysis. Afterwards, 32 key lncRNAs were screened using LASSO method (Figure 1B,C). The multivariate Cox regression analysis was utilized to determine independent prognostic factors (lncRNAs) of survival, and the results were visualized by a forest plot (Figure 2). As shown in Figure 2, five lncRNAs (AC015712.4, LINC02301, AGAP11, AC099850.3, and AC008915.1) may be the independent prognostic biomarkers in early-stage lung SCC (P value <0.05). Finally, the five lncRNAs were used to construct a risk assessment model to predict the survival outcome in patients with stage I and II lung SCC.
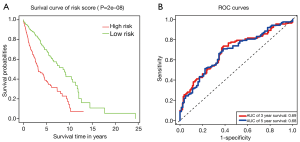
Evaluation of the 5-lncRNA-based risk model
According to the risk assessment model, all the 395 patients were classified into high‐risk and low‐risk groups, and patients in the low‐risk group had significantly better OS than those in the high‐risk group (Figure 3A). Moreover, the AUC values of ROC for the 5-lncRNA-based risk assessment model at 3- and 5-year OS were 0.69 and 0.68, respectively (Figure 3B). The results showed the 5-lncRNA-based risk assessment model had a general performance.
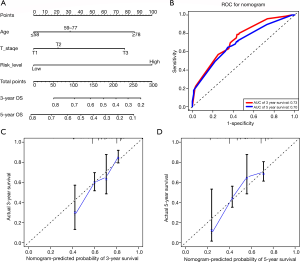
Establishment and assessment of the prognostic nomogram
X-Tile software was applied to determine the optimum threshold values for age. Then, we matched each sample with clinical information (including survival status, OS time, age, gender, TNM stage, T stage) according to patient barcode ID. In addition, the detailed clinical data and risk level of the 5-lncRNA-based risk assessment model of all 395 lung SCC patients were shown in Table 1.
Table 1
| Features | TCGA dataset (n=395) |
|---|---|
| Status (alive/dead) | 234/161 |
| Overall survival time, days (range/median) | 2–8,900/758 |
| Age (<58/59–77/≥78) | 61/293/41 |
| Gender (female/male) | 106/289 |
| TNM stage (I/II) | 239/156 |
| T stage (T1/T2/T3) | 107/250/38 |
| Risk level (high-risk/low-risk) | 197/198 |
The univariate and multivariate Cox regression analyses were conducted to assess independent prognostic factors. According to the results of univariate Cox regression analysis, the risk level, age, and T stage were significantly associated with OS in lung SCC patients (Table 2). In the multivariate analysis, risk level and T stage were still independent prognostic factors (Table 2). On the basis of the multivariate Cox regression analysis results, the nomogram that integrated independent prognostic factors (risk level, age, T stage) was established for predicting the 3- and 5-year OS probabilities in patients with stage I–II lung SCC (Figure 4A). Furthermore, AUC values were 0.73 (3-year ROC) and 0.70 (5-year ROC), which implied that the nomogram had a moderate performance (Figure 4B). The calibration curves also showed satisfactory agreement between nomogram predictions and actual observations in the probabilities of 3‐ (Figure 4C) and 5‐year OS (Figure 4D).
Table 2
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Risk level | |||||||
| Low | 1 (reference) | 1 (Reference) | |||||
| High | 2.496 | (1.798–3.464) | 4.6e-08* | 2.445 | (1.759–3.398) | 1.02e-07* | |
| Age (year) | |||||||
| ≤58 | 1 (reference) | 1 (Reference) | |||||
| 59–77 | 1.327 | (0.797–2.208) | 0.277 | 1.317 | (0.789–2.197) | 0.292 | |
| ≥78 | 2.232 | (1.175–4.242) | 0.014* | 2.142 | (1.124–4.082) | 0.021* | |
| Gender | |||||||
| Female | 1 (reference) | ||||||
| Male | 1.074 | (0.747–1.544) | 0.700 | – | – | – | |
| TNM stage | |||||||
| Stage I | 1 (reference) | ||||||
| Stage II | 1.257 | (0.910–1.735) | 0.166 | – | – | – | |
| T stage | |||||||
| T1 | 1 (reference) | 1 (Reference) | |||||
| T2 | 1.314 | (0.912–1.894) | 0.143 | 1.202 | (0.831–1.737) | 0.329 | |
| T3 | 2.032 | (1.112–3.714) | 0.021* | 1.988 | (1.082–3.652) | 0.027* | |
*, P<0.05. HR, hazard ratio; CI, confidence interval.
Results of KM survival analysis
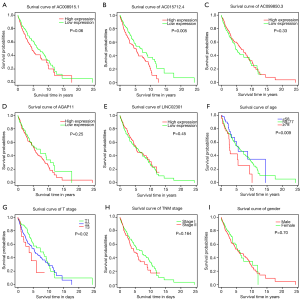
Results of KM survival analysis are displayed in Figure 5A,B,C,D,E,F,G,H,I. The survival time of stage I–II patients in the AC015712.4 high-expression group was poorer than that in the low-expression group (Figure 5B). Also, the oldest age group (≥78 years old) had a worse OS than the other two groups (Figure 5F). Besides, in the comparison of survival curves among the T1, T2, and T3 groups, the T3 group had worse OS than the other two groups (Figure 5G). None of the other comparisons were statistically significant.
Discussion
Even if early-stage NSCLC patients receive optimal treatments, survival rate remains low (22). Although lung adenocarcinoma and lung SCC belong to NSCLC, significant clinicopathologic differences exist between them (23). Furthermore, some cancer patients shared the same clinical and pathological stages, but their prognosis varied a lot. Thus, there is an urgent need to find effective prognostic biomarkers to predict the patient’s prognosis (24). Prior studies on the molecular mechanism of lung SCC have significantly improved the prognosis prediction for patients with lung SCC, but much of the literature on prognostic biomarkers is concerned with microRNAs or protein-coding genes (18,25,26).
In recent years, lncRNAs have been shown to be associated with prognosis in patients with lung cancer. For instance, previous studies have demonstrated that AFAP1-AS1, MNX1-AS1, and LINC01510 are associated with poor prognosis of NSCLC patients (27-29). In addition, He et al. developed an eight-gene prognostic signature, including seven mRNA and one lncRNA (SEC24B-AS1) which have great value in the prognosis of early-stage NSCLC (19). Moreover, recent studies have demonstrated that lncRNAs play an important role in lung SCC. Some lncRNA expression signatures in lung SCC have been found, which are associated with prognosis in lung SCC patients (9,18,20,30,31). However, the prognostic value of the lncRNA-based risk assessment model for predicting survival in stage I–II lung SCC patients still needs to be explored.
Nomogram is widely used to predict survival rates of cancer patients, and several nomograms for predicting survival of patients with lung SCC have been developed. For instance, Bi et al. built a prognostic nomogram to predict OS for patients with resected N2 stage lung SCC (32). Additionally, Zhu et al. established a nomogram model combining immune risk score and clinicopathologic parameter score to predict the OS at 3-year for patients with lung SCC (33). However, these studies of nomograms have not included variables related to lncRNAs.
In the present study, to construct the lncRNA-based risk assessment model for predicting the prognosis of patients with stage I–II lung SCC, univariate Cox regression and LASSO regression analyses were used to identify key lncRNAs. After that, the HR of each key lncRNA was obtained using multivariate Cox analysis. Moreover, all the patients were classified into low- and high-risk groups according to the risk assessment model, and the KM survival curve was conducted to estimate the association between the risk level and the survival rate. Furthermore, we construct a nomogram to easily predict the survival of patients with lung SCC. The prognostic power of the lncRNA-based risk assessment model and the nomogram were evaluated by ROC curves. Besides, KM survival curves were performed for several lncRNAs and clinicopathological factors.
A total of 2,021 DELs were found, of which 649 were downregulated and 1,372 were upregulated. In addition, 32 key lncRNAs were screened using Cox regression and LASSO analyses, which were then subjected to the multivariate Cox analysis. Five lncRNAs (AC015712.4, LINC02301, AGAP11, AC099850.3, and AC008915.1) were determined as independent prognostic biomarkers by multivariate analysis. Moreover, patients with stage I and II lung SCC in the high-risk group tended to have shortened survival time, and ROC curves indicated that the lncRNA-based risk assessment model is a feasible tool for prognostic prediction.
To predict the survival of patients with lung SCC easily, a prognostic nomogram integrated lncRNA-based risk assessment model and clinical factors (age and T stage) was constructed. Moreover, risk level of the lncRNA-based risk assessment model is the dominating independent prognostic factor. In addition, the ROC and calibration curves demonstrated that the nomogram was reliable in predicting OS rate. Although some lncRNAs did not reach the statistical significance in KM survival analysis, they also showed a trend of association with the prognosis of stage I and II lung SCC in the multivariate Cox analysis.
However, present research has some limitations. The five lncRNAs identified in the multivariate analysis have not been reported to be associated with NSCLC, and the biological function of these lncRNAs needs to be further validated with experiments. Besides, due to the limitations of the TCGA database, we could not obtain complete data regarding the administrations of chemotherapy, radiotherapy, and surgery. This lack of data may be the potential limitation of our prognostic nomogram.
In conclusion, we have constructed a 5-lncRNA-based risk assessment model which was significantly associated with the prognosis of patients with stage I–II lung SCC. Our data suggest that AC244502.3 may be a useful prognostic biomarker for lung SCC. Moreover, we also built a prognostic nomogram based on three variables (5-lncRNA-based risk assessment model, age, and T stage), which may help to improve the accuracy of clinical prognosis and outcome prediction of early-stage lung SCC. In the future, large-scale and multi-center studies are required to verify our findings.
Acknowledgments
We would like to thank the TCGA database for providing open access.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-999). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was not required, because this study was performed using a publicly accessible database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhou Y, Li S, Li J, et al. Effect of microRNA-135a on Cell Proliferation, Migration, Invasion, Apoptosis and Tumor Angiogenesis Through the IGF-1/PI3K/Akt Signaling Pathway in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2017;42:1431-46. [Crossref] [PubMed]
- Puri S, Shafique M, Gray JE. Immune Checkpoint Inhibitors in Early-Stage and Locally Advanced Non-Small Cell Lung Cancer. Curr Treat Options Oncol 2018;19:39. [Crossref] [PubMed]
- Zhang J, Fan J, Yin R, et al. A nomogram to predict overall survival of patients with early stage non-small cell lung cancer. J Thorac Dis 2019;11:5407-16. [Crossref] [PubMed]
- Asamura H, Goya T, Koshiishi Y, et al. A Japanese Lung Cancer Registry study: prognosis of 13,010 resected lung cancers. J Thorac Oncol 2008;3:46-52. [Crossref] [PubMed]
- Li Y, Gu J, Xu F, et al. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci Rep 2018;8:15834. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Liu H, Chen Y, Li Y, et al. miR195 suppresses metastasis and angiogenesis of squamous cell lung cancer by inhibiting the expression of VEGF. Mol Med Rep 2019;20:2625-32. [PubMed]
- Luo D, Deng B, Weng M, et al. A prognostic 4-lncRNA expression signature for lung squamous cell carcinoma. Artif Cells Nanomed Biotechnol 2018;46:1207-14. [Crossref] [PubMed]
- Lin T, Fu Y, Zhang X, et al. A seven-long noncoding RNA signature predicts overall survival for patients with early stage non-small cell lung cancer. Aging (Albany NY) 2018;10:2356-66. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120-4. [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci 2018;197:122-9. [Crossref] [PubMed]
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:443-9. [PubMed]
- Bo H, Fan L, Li J, et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer 2018;9:4677-83. [Crossref] [PubMed]
- Jing H, Qu X, Liu L, et al. A Novel Long Noncoding RNA (lncRNA), LL22NC03-N64E9.1, Promotes the Proliferation of Lung Cancer Cells and is a Potential Prognostic Molecular Biomarker for Lung Cancer. Med Sci Monit 2018;24:4317-23. [Crossref] [PubMed]
- Wang Y, Yang F, Zhuang Y. Identification of a progression-associated long non-coding RNA signature for predicting the prognosis of lung squamous cell carcinoma. Exp Ther Med 2018;15:1185-92. [PubMed]
- He R, Zuo S. A Robust 8-Gene Prognostic Signature for Early-Stage Non-small Cell Lung Cancer. Front Oncol 2019;9:693. [Crossref] [PubMed]
- Li S, Teng Y, Yuan MJ, et al. A seven long-noncoding RNA signature predicts prognosis of lung squamous cell carcinoma. Biomark Med 2020;14:53-63. [Crossref] [PubMed]
- Miao R, Ge C, Zhang X, et al. Combined eight-long noncoding RNA signature: a new risk score predicting prognosis in elderly non-small cell lung cancer patients. Aging (Albany NY) 2019;11:467-79. [Crossref] [PubMed]
- Bodelon C, Polley MY, Kemp TJ, et al. Circulating levels of immune and inflammatory markers and long versus short survival in early-stage lung cancer. Ann Oncol 2013;24:2073-9. [Crossref] [PubMed]
- Nakamura H, Sakai H, Kimura H, et al. Difference in Postsurgical Prognostic Factors between Lung Adenocarcinoma and Squamous Cell Carcinoma. Ann Thorac Cardiovasc Surg 2017;23:291-7. [Crossref] [PubMed]
- Wang P, Jin M, Sun CH, et al. A three-lncRNA expression signature predicts survival in head and neck squamous cell carcinoma (HNSCC). Biosci Rep 2018;38:BSR20181528. [Crossref] [PubMed]
- Chen B, Gao T, Yuan W, et al. Prognostic Value of Survival of MicroRNAs Signatures in Non-small Cell Lung Cancer. J Cancer 2019;10:5793-804. [Crossref] [PubMed]
- Zhu CQ, Strumpf D, Li CY, et al. Prognostic gene expression signature for squamous cell carcinoma of lung. Clin Cancer Res 2010;16:5038-47. [Crossref] [PubMed]
- Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer 2018;17:92. [Crossref] [PubMed]
- Li J, Wei L. Increased expression of LINC01510 predicts poor prognosis and promotes malignant progression in human non-small cell lung cancer. Biomed Pharmacother 2019;109:519-29. [Crossref] [PubMed]
- Liu G, Guo X, Zhang Y, et al. Expression and significance of LncRNA MNX1-AS1 in non-small cell lung cancer. Onco Targets Ther 2019;12:3129-38. [Crossref] [PubMed]
- Qi L, Zhang T, Yao Y, et al. Identification of lncRNAs associated with lung squamous cell carcinoma prognosis in the competitive endogenous RNA network. PeerJ 2019;7:e7727. [Crossref] [PubMed]
- Xiong Y, Zhang X, Lin Z, et al. SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1, and FEZF1-AS1 are crucial long non-coding RNAs associated with the prognosis of lung squamous cell carcinoma. Oncol Lett 2019;18:3985-93. [Crossref] [PubMed]
- Bi G, Lu T, Yao G, et al. The Prognostic Value Of Lymph Node Ratio In Patients With N2 Stage Lung Squamous Cell Carcinoma: A Nomogram And Heat Map Approach. Cancer Manag Res 2019;11:9427-37. [Crossref] [PubMed]
- Zhu Y, Zhang X. Investigating the significance of tumor-infiltrating immune cells for the prognosis of lung squamous cell carcinoma. PeerJ 2019;7:e7918. [Crossref] [PubMed]


