
Protein tyrosine phosphatase receptor type delta (PTPRD) suppresses the expression of PD-L1 in human hepatocellular carcinoma by down-regulating STAT3
Introduction
Hepatocellular carcinoma (HCC) is one of the most common tumors that threaten human life and health around the world. According to global cancer statistics from 2018, there were 841,080 newly diagnosed HCC patients and 781,631 deaths worldwide (1). HCC is a disease with an extremely high degree of malignancy and an abysmal prognosis. Although great advancements in early detection and surgical treatment of HCC have been made globally, the prognosis of most patients with HCC is still relatively poor due to it having high rates of recurrence and metastasis and a 5-year survival rate of less than 12% (2). Many factors, including cirrhosis, distant metastasis, and immune escape, contribute to the high mortality rate in HCC patients (3). There is also increasing evidence that immune escape is a critical factor that contributes to the development and progression of HCC (4,5). However, the immune escape pathways of HCC are complex, involving perturbations of antigen presentation, immune effector functions, the disarray of cytokine profiles, and alterations of immune checkpoint molecules (2). Programmed cell death (PD-1) and programmed cell death ligand 1 (PD-L1) play an essential role in immune checkpoints among these mechanisms. Therefore, it is urgent that the molecular mechanisms of PD-L1 in advanced HCC are understood and new therapeutic strategies are developed.
Protein tyrosine phosphatase receptor type delta (PTPRD) is a member of a family of protein tyrosine phosphatases (PTPs), and its functions include the regulation of cell growth and differentiation (6). PTPRD is often inactivated in many human cancers due to deletions or epigenetic mechanisms. Furthermore, it takes part in the signal transduction of tumor cells (7). Previous studies have reported that PPTRD deficiency occurs in multiple types of human malignancies, including HCC (8), urothelial carcinomas (9), neuroblastoma (10), esophageal cancer (11), and lung adenocarcinoma (12). Acun et al. found that PTPRD expression levels were low or even undetectable in 11 out of 14 cell lines. Additionally, their results showed that 82.6% of primary HCCs in their clinical samples displayed low PTPRD expression (8). Another study showed that deletions of PTPRD were also present in rat hepatomas and the human hepatoblastoma HepG2 cell line, which demonstrates that PTPRD deletions contribute to the development of the malignant phenotype in hepatocytes (13) .
Signal transducer and activator of transcription 3 (STAT3) is known as a major transcription factor involved in many cellular processes. Some of the processes in which STAT3 plays a role are cell growth and proliferation, differentiation, migration, and cell death or cell apoptosis (14). A recent study has shown that STAT3 could positively regulate PD-L1 expression to promote immune escape in human cancer (15). Loss of PTPRD can induce abnormal activation of STAT3, promoting proliferation, differentiation of tumor cells, and inhibiting immune activity (7). Also, in the study of glioblastoma multiforme, researchers showed that phosphorylated STAT3 (p-STAT3) is a substrate of PTPRD. These results demonstrate that allelic loss of PTPRD causes p-Stat3 accumulation and Stat3 hyperactivation, eventually leading to the promotion of gliomagenesis (16). However, whether PTPRD is correlated with PD-L1 expression has not been reported or confirmed.
In the present study, we aimed to describe the relationship between PTPRD and PD-L1 in HCC cells and elucidate its potential molecular mechanisms. Here, we found that PTPRD was significantly down-regulated in HCC tissues compared to the matched adjacent tissues, while the opposite results were found for PD-L1. Also, our results show that PTPRD may suppress PD-L1 expression in HCC cells through negative regulation of STAT3.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2425).
Methods
Antibodies and reagents
Human HCC cell lines HepG2 and Huh-7 were obtained from the Meixuan Company (Shanghai, China) and the Cell Bank of the Chinese Academic of Sciences (Shanghai, China). Normal hepatocyte line L02 was obtained from Procell (Wuhan, China). All cells were cultured in DMEM (C11995500BT, Gibco, USA) supplemented with 10% fetal bovine serum (900–108, Gemini) and 1% penicillin/streptomycin (Gibco, USA) at 37 °C with 5% CO2. The PTPRD overexpression adenovirus and knockdown lentivirus were bought from Genechem Co., Ltd. (Shanghai, China). The antibodies against PD-L1 (ab205921), PTPRD (ab103013), STAT3 (ab32500) and p-STAT3 (phosphor Y705, ab76315) were bought from Abcam (Cambridge, UK).
Patients and tissue sample collection
Sixteen tumorous and 16 adjacent normal tissues (16 pairs) from HCC patients who received surgical resections at the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China). All patients signed the informed consent form after being informed of their data being used for research purposes. None of the individuals had ever received any treatment before. The usage of tissues in this study was authorized by the ethics committee of the Guangxi Medical University. This study was conducted in adherence with the guidelines of the Declaration of Helsinki (as revised in 2013).
Quantitative real-time polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from the clinical specimens and cell lines using Trizol reagent, following the manufacturer’s instructions (Takara, Japan). Reverse transcription was conducted using the PrimeScript RT Regent Kit (Takara, Japan) following the manufacturer’s guidelines. Real-time PCR was conducted on the ABI 7500 Taqman System (Applied Biosystems, Foster, CA, USA) using the SYBR Green Real-Time PCR Master Mix Kit and specific primers, which were designed by Invitrogen (Carlsbad, USA). The primer sequences used were as follows: PTPRD, forward 5'-TTTACACGAACACCCGTTGA-3, and reverse 5'-CGGAGTCCGTAAGGGTTGTA-3; PD-L1, forward 5'- TGTGGCATCCAAGATACAAACTCAAAG-3,and reverse 5'-TCCTCCTCTGCTTTCGCCAGGTTC-3; GAPDH, forward, 5'-CAGCCTCAAGATCATC AGCA-3, and reverse 5'-TGTGGTCATGAG TCCTTCCA-3. The melt curves were used to analyze the accuracy of the PCR reaction. The 2−ΔΔCt values were calculated to evaluate the expressions of genes.
Western blotting
Proteins were extracted from the HCC samples and human HCC cells, respectively, using a RIPA lysis buffer (Beyotime, Shanghai, China) containing 1 nM PMSF (Invitrogen), following the manufacturer’s instructions. Next, the lysate was centrifuged at 120,000 rpm to remove the insoluble materials. The protein concentrations were qualified using a bicinchoninic acid protein assay kit (Thermo Fisher, Waltham, MA, USA). Subsequently, 20 µg of protein per well was electrophoresed in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and then transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA) and incubated with primary antibodies at 4 °C overnight. The membrane was then incubated with horseradish peroxidase (HRP)-labeled secondary antibody at xyz for 2 h. The bands were detected using an ECL Western Blotting Kit (Millipore, Temecula, CA, USA) and analyzed on the Alpha gel imaging system (Protein Simple, Silicon Valley, USA).
Cell transfection
HepG2 cells were seeded at a density of 100,000 cells per well on a 6-well culture plate. Then, the cells were incubated until the culture reached 60–80% confluence. After that, the cells were transfected with either the PTPRD empty vector, PTPRD overexpression adenovirus, or the lentiviral expression system for PTPRD shRNA (Genechem Shanghai, China) in HepG2 cells, using the Transfection Reagent (Genechem Shanghai, China), following the manufacturer’s instructions. After 48 h of transfection, the total RNA and protein levels were isolated from the cells. Transfection efficiency was determined with RT-PCR and Western blotting.
Statistical analysis
All data were expressed as the mean ± standard deviation. SPSS Version 22.0 software (IBM Corp., Armonk, NY, USA) was used to perform the data analysis. Differences between two groups were analyzed using Student’s t-test. Differences between three or more groups were analyzed by a one-way ANOVA. Following that, the relationship between PTPRD expression and PD-L1 expression in HCC tissues was evaluated with Spearman’s correlation analysis. Significance was represented by P values of <0.05.
Results
The mRNA and protein expressions of PTPRD in HCC specimens and normal adjacent tissues
The mRNA and protein expressions of PTPRD in fresh HCC specimens and normal adjacent tissues were investigated to evaluate the expression of PTPRD in HCC. RT-PCR and western blotting analysis demonstrated that the mRNA and protein expressions of PTPRD were significantly decreased in the HCC samples compared with their normal adjacent tissue counterparts (P<0.01), as shown in Figure 1.
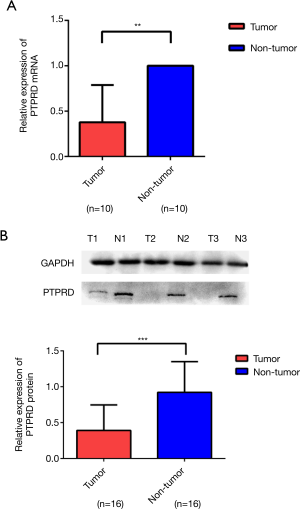
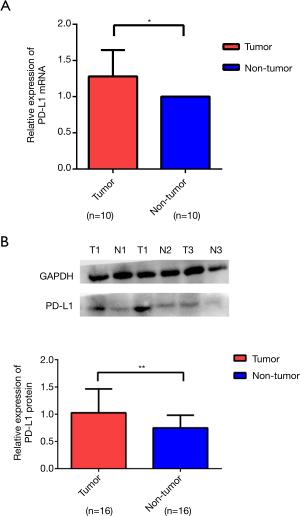
The mRNA and protein expressions of PD-L1 in HCC specimens and normal adjacent tissues
We used RT-PCR and western blotting to analyze the expression levels of PD-L1 in tissue samples from the same patient. The expression levels of PD-L1 were significantly increased in cancer tissues compared with those in the adjacent normal tissues (Figure 2A,B).
Relationships between PTPRD or PD-L1 expression and patients clinicopathological features
Pearson’s correlation analysis was carried out to investigate the associations between the protein expressions of PTPRD and PD-L1. The results showed that PTPRD protein expression was negatively correlated with PD-L1 protein expression in the HCC tissues (Figure 3A). However, we did not find a significant correlation between PTPRD expression and PD-L1 in the para-carcinoma samples (Figure 3B). These results showed that there is a complex regulatory effect in clinical tumors. Then, we summarized the correlations between the clinicopathological features of the patients and PTPRD expression (Table 1). We found that the lower the expression level of PTPRD, the more likely the occurrence of hepatitis B (P=0.045) and the higher expression levels of ALT (P=0.016) (Table 1). However, there were no significant correlations between PTPRD expression and other clinicopathological parameters, such as age and tumor size. Interestingly, we found that PD-L1 protein expression was also significantly associated with the presence of hepatitis B (Table 1). Further, we used a T-test to analyze the expression of PTPRD and PD-L1 and the clinicopathological features of each layer tissue. We found that they were significantly expressed in most of the pathological features.
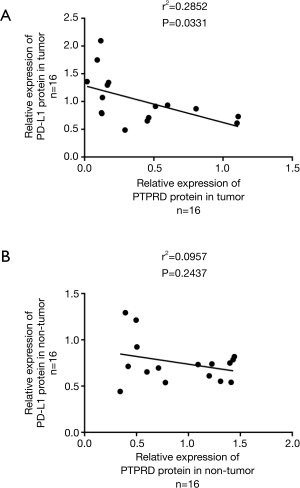
Table 1
| Clinicopathological feature | n | Relative expression of PTPRD protein | Pa | Relative expression of PD-L1 protein | Pa | Pb |
|---|---|---|---|---|---|---|
| Age, years | 0.282 | 0.731 | ||||
| ≤50 | 8 | 0.2923±0.1900 | 1.0647±0.5049 | 0.010* | ||
| >50 | 8 | 0.4924±0.4583 | 1.0718±0.3861 | 0.037* | ||
| HBsAg | 0.350 | 0.661 | ||||
| Negative | 3 | 0.5711±0.5119 | 1.1303±0.5429 | 0.264 | ||
| Positive | 13 | 0.3512±0.3215 | 1.0007±0.4346 | 0.000** | ||
| Hepatitis B | 0.045* | 0.019* | ||||
| Negative | 5 | 0.7403±0.4015 | 0.7406±0.1201 | 0.999 | ||
| Positive | 11 | 0.2343±0.1891 | 1.1543±0.4744 | 0.000** | ||
| AFP, ng/mL | 0.997 | 0.192 | ||||
| ≤25 | 5 | 0.3920±0.4373 | 1.3114±0.5826 | 0.022* | ||
| >25 | 11 | 0.3924±0.3345 | 0.8948±0.3070 | 0.002** | ||
| ALT, U/L | 0.016* | 0.939 | ||||
| ≤40 | 3 | 0.1556±0.0234 | 1.0067±0.3274 | 0.011* | ||
| >40 | 13 | 0.4469±0.3737 | 1.0292±0.4725 | 0.011* | ||
| Tumor size, cm | 0.617 | 0.532 | ||||
| ≤5 | 5 | 0.3119±0.1996 | 1.1174±0.6108 | 0.004** | ||
| >5 | 11 | 0.4192±0.3964 | 0.9592±0.3796 | 0.008** | ||
| BCLC stage | 0.127 | 0.761 | ||||
| A | 10 | 0.4996±0.3934 | 0.9978±0.4794 | 0.031* | ||
| C | 6 | 0.2191±0.1874 | 1.0703±0.4019 | 0.002** |
Pa value represents the significant of PTPRD or PD-L1 expressions in different levels of clinicopathological features; Pb value represents the significant of PTPRD and PD-L1 expressions in the same level of clinicopathological features. *, P<0.05; **, P<0.01 were considered statistically significant. PTPRD, protein tyrosine phosphatase receptor type delta; PD-L1, programmed cell death ligand 1; BCLC; Barcelona Clinic Liver Cancer; AFP, alpha fetoprotein.
The mRNA and protein expressions of PTPRD and PD-L1 in HepG2 cells
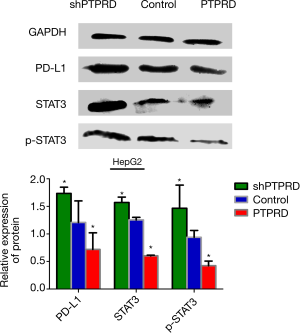
The mRNA and protein expressions of PTPRD in HepG2 and Huh-7 were significantly reduced compared to the normal hepatocyte line L02 (Figure 4A), it is very helpful for us to further study the effect of PTPRD in HCC cells. We transfected them with either the PTPRD empty vector, PTPRD overexpression adenovirus, or the lentiviral expression system for PTPRD shRNA in the HepG2 cells after affirming the expression of PTPRD in HCC cells. The transfection efficiency was measured through RT-PCR and western blot analysis (Figure 4B,C). As shown in Figure 4D,E, compared with the control group, over-expression of PTPRD significantly inhibited the expression of PD-L1 in HepG2 cells, while PTPRD ablation significantly increased PD-L1 expression in HepG2 cells.
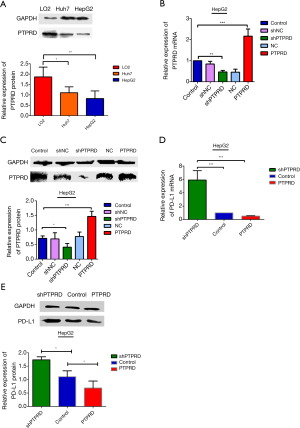
PTPRD modulated PD-L1 expression via STAT3 in HepG2 cells
We further detected the expression levels of STAT3 and p-stat3 proteins in the HepG2 cells with PTPRD overexpression or knockdown to understand the molecular mechanisms of the PTPRD effects the expression on PD-L1 in HCC cells. The phosphorylation of STAT3 decreased in the PTPRD-overexpression cells and increased in the PTPRD knockdown cells (Figure 5), which was consistent with the changes in PD-L1 protein expression levels with a statistically significant difference. These results suggested that PTPRD affects the expression of PD-L1 via the STAT3 pathway.
Discussion
HCC is one of the most common malignancies in the world. It ranks among the top 10 cancers for morbidity and mortality and poses a great threat to public health. Currently, chemotherapy, radiotherapy, targeted therapy, and surgical resection are the main therapeutic methods for HCC (17). In recent years, great progress has been made on the treatment outcomes of HCC; however, the long-term prognosis is still disappointing for HCC patients. Therefore, there is an urgent need to discover new therapies that will improve the prognosis of patients with advanced HCC. Tumor immunotherapy has one of the highest potentials for new developments. Therefore, it is vital to find the key genes which are implicated in tumor evasion from immunosurveillance. As a vital member of the B7 immunoglobulin superfamily, PD-L1 is a type I transmembrane glycoprotein with a molecular weight of 50 kD (18). The binding of the ligand PD-L1 to its receptor PD-1 transduces a negative signal into T-cells, inhibiting their activation, thereby leading to tumor immune escape (19). In HCC, the interaction between PD-L1 and PD-1 impairs effector T-cell function, and in vitro disruptions of this interaction restore the function of the tumor-derived effector T-cells (20). A large number of studies have shown that PD-L1 is overexpressed in most solid tumor tissues such as HCC, non-small cell lung cancer, gastric cancer, and esophageal cancer. Meanwhile, PD-L1 expression is positively associated with tumor size, vascular invasion, and tumor stage classification (21-24). It has been suggested that PD-L1 may play an essential role in tumor development. However, the PD-L1 overexpression role in HCC is still poorly understood.
The PTPRD gene is members of the PTP family. It has been reported (25) that PTPRD acts as a tumor suppressor gene, and it plays a crucial role in controlling cell proliferation, apoptosis, survival, immunity, migration, and invasion. Earlier studies showed that PTPRD and PD-L1 are implicated in multiple types of human malignant tumors. Nonetheless, their relationship in cancers is still poorly understood. Our study identified that PTPRD was significantly down-regulated in cancerous tissues compared to para-cancerous tissues; however, PD-L1 was significantly overexpressed in cancerous tissues compared with in para-cancerous tissues. One-way ANOVA tests showed that a lower PTPRD level was associated with higher hepatitis B virus (HBV) and ALT rates. Additionally, it is worth mentioning that PD-L1 expression was also related to Hepatitis B. However, there was no association between PTPRD or PD-L1 expression and other clinical parameters, such as age, AFP, and BCLC stage. It has been reported that HBV infection is a major factor in promoting the development and progression of HCC, and chronic viral infection and tumor microenvironments can severely impair the proliferative capacity and effector functions of immune cells, rendering the immune cells such as T cell unable to eliminate the virus or to reject the tumors (26). Thus, we hypothesized that the expressions of PTPRD and PD-L1 might be related to the pathogenesis of hepatitis B-induced HCC, and the loss of PTPRD may be linked to immune response.
Interestingly, we used a t-test to analyze the expression of PTPRD and PD-L1 in the clinicopathological features of each patient and found that they were significantly expressed in most of the pathological features. Also, our study is the first to show a negative correlation between PTPRD and PD-L1 in HCC tissues. The finding provides a negative regulation pattern of PD-L1 in HCC, and the regulation of PTPRD activity would be important for the expression of PD-L1. However, only 16 patients with HCC were included in this study. There is still controversy surrounding the median by which high and low PTPRD and PD-L1 expressions are classified. These factors may have affected the results of our experiment. The overexpression of PD-L1 is mainly driven by the transcriptional regulation of the signaling pathways. STAT3 as an important transcriptional factor has been identified as a critical regulator of PD-L1 expression in various cell systems. Such as PD-L1 is transcriptionally regulated by STAT3 in cell lymphoma and the STAT3 pathway regulates the expression of PD-L1 on non-small lung cancers (27,28).
Based on our preliminary results, we hypothesized that PTPRD might be involved in the regulation of PD-L1 expression in HCC. We performed further studies to confirm our hypothesis and explore the relevant mechanism. We used gain- and loss-of-function approaches to manipulate PTPRD expression in the HCC cells. Over-expression and knockdown investigations were conducted on HepG2 cells. Our results showed that overexpression of PTPRD inhibited PD-L1 expression in HCC cells, while knockdown of PTPRD promoted PD-L1 expression in HCC cells. Next, we evaluated PTPRD the intracellular signaling pathways dictating tumor PD-L1 expression. Activation of the STAT3 pathways has been reported to drive PD-L1 expression, STAT3 is a critical regulator of PD-L1 expression in various cell systems (29,30). Van Renne et al. confirmed STAT3 to be a target for PTPRD and play a crucial role in liver regeneration; there was a significant correlation between PTPRD levels and STAT3 transcriptional activities in the livers of patients (31). Changes in STAT3 and p-stat3 proteins were detected in HepG2 cells with PTPRD overexpression or knockdown. The results showed that STAT3 and p-stat3 proteins decreased significantly when PTPRD was overexpressed. Meanwhile, when PTPRD was knocked down, the protein levels of STAT3 and p-stat3 significantly increased. Furthermore, the up-regulation and downregulation of STAT3 were consistent with PD-L1. Therefore, we hypothesized that PTPRD repressed PD-L1-mediated immune escape of HCC cells may through the down-regulation of STAT3. Whether this is an important negative regulation mechanism for STAT3 is worth further study. Recent years, PTPRD has been studied as a tumor suppressor gene and has now become an attractive target for cancer therapy. Our study suggests that the overexpression of PTPRD in HCC inhibitor the expression of PD-L1. Whether this effect will lead to cancer evasion during depletion with PTPRD should be evaluated.
Our research had some limitations. The small sample size of patients who were included in the study is the largest limitation. Further, our sample lacks stage IV patients, and the patient information was not complete enough. Additionally, the mechanisms of PTPRD/PD-L1 signaling and its impact on tumor progression requires further basic and clinical research. Therefore, there may have been some bias in our results.
In conclusion, our study is the first to show that PTPRD and PD-L1 are negatively correlated in HCC tissues, and low expression of PTPRD and overexpression of PD-L1 were related to HBV. Also, our findings revealed that PTPRD inhibited the expression of PD-L1 in HCC cells via the STAT3/PD-L1 axis. Therefore, this work supplies new insights into the understanding of the underlying molecular mechanisms of HCC and suggests that targeting the PTPRD /PD-L1 axis could be a potential therapeutic choice for HCC patients.
Acknowledgments: The authors thank Min Dong for providing help and support. Min Dong initiated and coordinated the whole study, participated in the design of the study, drafting the manuscript and data interpretation. All authors have read and approved the final manuscript. This work was supported by National Natural Science Foundation of China and Guangxi Natural Science Foundation.
Acknowledgments
Funding:
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2425
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2425
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2425). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in adherence with the guidelines of the Declaration of Helsinki (as revised in 2013) and received approval from the Ethics Committee of Guangxi University. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Nenu I, Breaban I, Pascalau S, et al. The future is now: beyond first line systemic therapy in hepatocellular carcinoma. Transl Cancer Res 2019;8:S261-74. [Crossref]
- Guo J, Li L, Guo B, et al. Mechanisms of resistance to chemotherapy and radiotherapy in hepatocellular carcinoma. Transl Cancer Res 2018;7:765-81. [Crossref]
- Li W, Wang H, Ma Z, et al. Multi-omics Analysis of Microenvironment Characteristics and Immune Escape Mechanisms of Hepatocellular Carcinoma. Front Oncol 2019;9:1019. [Crossref] [PubMed]
- Zhu HF, Liu YP, Liu DL, et al. Role of TGFbeta3-Smads-Sp1 axis in DcR3-mediated immune escape of hepatocellular carcinoma. Oncogenesis 2019;8:43. [Crossref] [PubMed]
- Du Y, Grandis JR. Receptor-type protein tyrosine phosphatases in cancer. Chin J Cancer 2015;34:61-9. [Crossref] [PubMed]
- Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer 2006;6:307-20. [Crossref] [PubMed]
- Acun T, Demir K, Oztas E, et al. PTPRD is homozygously deleted and epigenetically downregulated in human hepatocellular carcinomas. OMICS 2015;19:220-9. [Crossref] [PubMed]
- Beothe T, Zubakov D, Kovacs G. Homozygous losses detected by array comparative genomic hybridization in multiplex urothelial carcinomas of the bladder. Cancer Genet 2015;208:434-40. [Crossref] [PubMed]
- Stallings RL, Nair P, Maris JM, et al. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res 2006;66:3673-80. [Crossref] [PubMed]
- Frankel A, Armour N, Nancarrow D, et al. Genome-wide analysis of esophageal adenocarcinoma yields specific copy number aberrations that correlate with prognosis. Genes Chromosomes Cancer 2014;53:324-38. [Crossref] [PubMed]
- Ren Y, Su H, She Y, et al. Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules. Transl Lung Cancer Res 2019;8:235-46. [Crossref] [PubMed]
- Nalesnik MA, Tseng G, Ding Y, et al. Gene deletions and amplifications in human hepatocellular carcinomas: correlation with hepatocyte growth regulation. Am J Pathol 2012;180:1495-508. [Crossref] [PubMed]
- McFarland BC, Benveniste EN. Reactive astrocytes foster brain metastases via STAT3 signaling. Ann Transl Med 2019;7:S83. [Crossref] [PubMed]
- Chen J, Jiang CC, Jin L, et al. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol 2016;27:409-16. [Crossref] [PubMed]
- Ortiz B, Fabius AW, Wu WH, et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci U S A 2014;111:8149-54. [Crossref] [PubMed]
- Ho V, Lim TS, Lee J, et al. TLR3 agonist and Sorafenib combinatorial therapy promotes immune activation and controls hepatocellular carcinoma progression. Oncotarget 2015;6:27252-66. [Crossref] [PubMed]
- Shi L, Chen S, Yang L, et al. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J Hematol Oncol 2013;6:74. [Crossref] [PubMed]
- Xie F, Xu M, Lu J, et al. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer 2019;18:146. [Crossref] [PubMed]
- Zhou G, Sprengers D, Boor PPC, et al. Antibodies Against Immune Checkpoint Molecules Restore Functions of Tumor-Infiltrating T Cells in Hepatocellular Carcinomas. Gastroenterology 2017;153:1107-19.e10. [Crossref] [PubMed]
- Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res 2005;11:2947-53. [Crossref] [PubMed]
- Daum O, Dubová M, Skálová A, et al. Predictive diagnostics of gastric cancer in 2018. Cesk Patol 2018;54:23-6. [PubMed]
- Sun X, Roudi R, Chen S, et al. Immune-related adverse events associated with PD-1 and PD-L1 inhibitors for nonsmall cell lung cancer: Protocol for a systematic review and meta-analysis. Medicine (Baltimore) 2017;96:e8407. [Crossref] [PubMed]
- Li Z, Li B, Peng D, et al. Expression and clinical significance of PD-1 in hepatocellular carcinoma tissues detected by a novel mouse anti-human PD-1 monoclonal antibody. Int J Oncol 2018;52:2079-92. [Crossref] [PubMed]
- Chan TA, Heguy A. The protein tyrosine phosphatase receptor D, a broadly inactivated tumor suppressor regulating STAT function. Cell Cycle 2009;8:3063-4. [Crossref] [PubMed]
- Sun C, Lan P, Han Q, et al. Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun 2018;9:1241. [Crossref] [PubMed]
- Abdelhamed S, Ogura K, Yokoyama S, et al. AKT-STAT3 Pathway as a Downstream Target of EGFR Signaling to Regulate PD-L1 Expression on NSCLC cells. J Cancer 2016;7:1579-86. [Crossref] [PubMed]
- Atsaves V, Tsesmetzis N, Chioureas D, et al. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia 2017;31:1633-7. [Crossref] [PubMed]
- Mimura K, Teh JL, Okayama H, et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci 2018;109:43-53. [Crossref] [PubMed]
- Ding L, Chen X, Xu X, et al. PARP1 Suppresses the Transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol Res 2019;7:136-49. [Crossref] [PubMed]
- Van Renne N, Roca Suarez AA, Duong FHT, et al. miR-135a-5p-mediated downregulation of protein tyrosine phosphatase receptor delta is a candidate driver of HCV-associated hepatocarcinogenesis. Gut 2018;67:953-62. [Crossref] [PubMed]
(English Language Editors: J. Chapnick and J. Reynolds)


