
Expression of biogenesis of ribosomes BRX1 is associated with malignant progression and prognosis in colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common cancers in China. Both the incidence and mortality of CRC ranked fifth in 2015, with 276,000 new cases and 191,000 deaths, respectively (1). Surgical resection, chemotherapy, and molecular targeted therapy are the major treatment options for CRC. However, 30–40% of patients who receive systematic treatment will relapse (2). The 5-year survival rate of CRC in China is around 55%, and the 5-year survival rate of patients with stage IV CRC is <10% (3). It is important that the therapeutic regime is further improved to achieve a higher survival rate in patients.
Despite the rapid development of molecular targeted drugs in recent years, only several drugs, such as bevacizumab, cetuximab, and panitumumab, are currently recognized as molecular targeted drugs for CRC (4,5). According to the 2019 National Comprehensive Cancer Network guidelines for CRC, these three molecular targeted drugs are used as supplements for general chemotherapy programs and are only recommended for use in patients with the KRAS wild type (6). It has been suggested that these molecular targeted drugs cannot be dependently used for the treatment of CRC patients. The main limitation of current molecular targeted drugs is that they are targeted at only one or one kind of protein, whereas the expression of the specific protein in cancer cells is different due to their different progression and development. There are two solutions to this problem; one is to increase the type of drug targets, and the other is to find new targets to improve the inhibitory effects of tumors through interactions between different targets (7). The former has high risks due to the side-effects of drugs. The latter is the currently preferred option, because it can inhibit the growth of tumors more safely and effectively.
The ribosome synthesis superfamily plays an important role in the process of ribosome synthesis and rRNA binding, which can affect cell proliferation and differentiation (8). Growing evidence indicates that upregulated ribosome biogenesis is associated with an increased risk of cancer onset (9). The biogenesis of ribosomes BRX1 (BRIX1) gene, which is involved in the synthesis of ribosomal 60S subunits, is located in cytoband 5p13.2 and is expressed at highest in pulmonary alveoli (10). It has been reported that its expression is related to smoking habits and myocardial damage due to smoking and PM2.5 exposure (11,12). Studies on BRIX1 in cancer are rare. Jung et al. reported the significantly increased expression of BRIX1 in a variety of tumors (13). However, the relationship between the expression of BRIX1and the progression and prognosis of CRC is still unclear. In the present prospective study, we measured the expression of BRIX1 in patients with CRC to determine the role of BRIX1 in the progression of CRC and provide preliminary evidence for the development of novel molecular targeted drugs for CRC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2564).
Methods
Patients and specimens
Patients who received surgical treatment for CRC between January 2018 and December 2018 at the Affiliated Nanjing Jiangbei Hospital of Nantong University were enrolled. In the present study, CRC was defined as tumors located between the ileocecal valve and dentate line. The following were excluded: (I) patients <18 years of age; (II) patients who refused to sign an informed consent form; (III) patients who were diagnosed with metastatic CRC; (IV) patients who had severe organ dysfunction; (V) patients who had life-threatening postoperative complications; and (VI) patients who had previously received surgical treatment for CRC. CRC tissue samples and adjacent normal tissue samples from patients were collected. Collected tissues were used for further quantitative reverse transcription–polymerase chain reaction (RT-PCR) and Western blot. All enrolled patients were followed up for 12 months. The outcome in the present study was overall survival of enrolled patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Affiliated Nanjing Jiangbei Hospital of Nantong University (No. J2017034), and informed consent was taken from all the patients.
Data collection
The following patient characteristics at admission were collected: age, sex, anatomical location, tumor stage, differentiation stage, and the level of carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA199). Anatomical location was defined as right-sided colon, left-sided colon, and rectum. Tumor stage was defined as I, II, III, or IV stage, according to TNM stage. Differentiation stage was defined as well, moderate, or poor. The CEA level was defined as elevated (>5 ng/L) or normal (≤5 ng/L), and the CA199 level was defined as elevated (>37.00 µ/mL) or normal (≤37.00 µ/mL).
RNA isolation
The total RNA of collected tissues was extracted using a total RNA extraction kit (Solarbio). The total RNA was quantified using the qubit flex fluorometer (ThermoFisher Scientific) at 260 nm. The total RNA was then reverse-transcribed using the PrimeScript RT reagent kit (Takara Bio), according to the manufacturer’s instructions.
Quantitative RT-PCR
The mRNA levels of BRIX1 genes were quantified by the ABI PRISM 7500 sequence detection system (Applied Biosystems). The primer pair used in the present study was as follows: forward: CCCGTCTTCCAGTCTCCTAA; reverse: TGTCCAGGGTTTCCTTCG. All assays in the present study were performed in three duplicated repeats. The data were automatically collected and analyzed by SDS version 1.3.1 (Applied Biosystems).
Protein extraction
The protein in the tissue sample was extracted using a total protein extraction kit (Solarbio). Briefly, 100 mg tissue was washed with phosphate buffer saline and minced. The minced tissue was then immersed in lysis buffer for 2–4 hours and centrifuged at 12,000 ×g for 30 minutes at 4 °C. The supernatant was collected and the protein was quantified using a bicinchoninic acid protein assay kit (ThermoFisher Scientific).
Western blot
In order to perform the Western blot assay, 20 mg protein was separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% bovine serum albumin and incubated overnight with primary antibodies against BRIX1 (Sino Biological). The membranes were then incubated with secondary antibodies for 1 hour at room temperature. Glyceraldehyde-3-phosphate dehydrogenase was used as the reference. The blot was quantified using Image J software version 1.51 (National Institutes of Health).
Statistical analysis
Continuous variables in the present study were reported as mean ± standard deviation, and other categorical variables were reported as numbers with percentages. The differences of continuous variables and categorical variables between the two groups were analyzed using Mann-Whitney U-test and χ2 test, respectively. GraphPad Prism 7 software (GraphPad Software) was used for the Kaplan-Meier survival curve, and the differences between the two groups were analyzed using log-rank test. Moreover, the Cox proportional hazards model was used to analyze the risk factors of overall survival in CRC patients. Differences were considered to be significant if P<0.05. The statistical analysis was performed using Stata16.0 (StataCorp LLC).
Results
Patient characteristics
Sixty CRC patients were enrolled in the present study. Their characteristics are shown in Table 1. There were 38 male patients and 22 female patients in the study. The mean age of the enrolled patients was 60.4±9.6 years. The tumors in 23, 15, and 22 patients were located at the right-sided colon, left-sided colon, and rectum, respectively. CRC patients at all stages were enrolled in the present study; 9 patients were stage I, 24 were stage II, 16 were stage III, and 11 were stage IV. Differentiation stage data were collected according to histopathological examinations. As a result, 16 tumors were found to be well differentiated, 31 were moderately differentiated, and 13 were poorly differentiated. The CEA and CA199 levels were also recorded. As seen in Table 1, the CEA level was elevated in 41 patients, and the CA199 level was elevated in 29 patients.
Table 1
| Characteristics | Total |
|---|---|
| N | 60 |
| Sex | |
| Male | 38 |
| Female | 22 |
| Age (years) | 60.4±9.6 |
| Anatomical location | |
| Right-sided colon | 23 |
| Left-sided colon | 15 |
| Rectum | 22 |
| Tumor stage | |
| I | 9 |
| II | 24 |
| III | 16 |
| IV | 11 |
| Differentiation stage | |
| Well-differentiated | 16 |
| Moderately differentiated | 31 |
| Poorly differentiated | 13 |
| CEA | |
| Normal | 19 |
| Elevated | 41 |
| CA199 | |
| Normal | 31 |
| Elevated | 29 |
CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen.
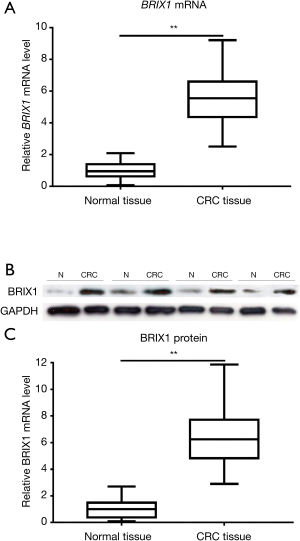
BRIX1 level was significantly higher in CRC tissues
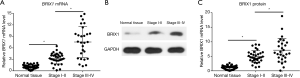
The BRIX1 mRNA level in CRC tissues and adjacent normal tissues was measured using quantitative RT-PCR. As seen in Figure 1A, the BRIX1 mRNA level in CRC tissues was higher than that in adjacent normal tissues (1.0±0.5 vs. 5.5±1.7, P<0.01). We then measured the BRIX1 protein level in both tissues. A representative diagram of the western blot is shown in Figure 1B. The BRIX1 protein level in CRC tissues was significantly higher than that in adjacent normal tissues (1.0±0.6 vs. 6.4±2.1, P<0.01) (Figure 1C). Furthermore, we divided all enrolled patients into two groups: stages I–II and stages III–IV. Stage III–IV patients were found to have a dramatically higher level of BRIX1 mRNA and protein compared with stage I–II patients (Figure 2A,B,C; P<0.05).
BRIX1 level was related to the prognosis of CRC patients
The enrolled patients were divided into two groups based on the relative level of the BRIX1 protein in CRC tissues (>6 or ≤6). The Kaplan-Meier survival curve was used to compare the difference in the overall survival rate between the two groups. As seen in Figure 3, the overall survival rate of patients with a high level of the BRIX1 protein was much lower than that of patients with a low level of the BRIX1 protein (P=0.022). The Cox proportional hazards model was further used to analyze the risk factors of overall survival in CRC patients. As shown in Table 2, three variables acted as risk factors of overall survival in CRC patients according to the multivariate analysis: tumor stage III or IV [P=0.044, hazards ratio (HR) =6.711], poor differentiation (P=0.022, HR =5.423), and elevated expression of the BRIX1 protein (P=0.048, HR =4.988).
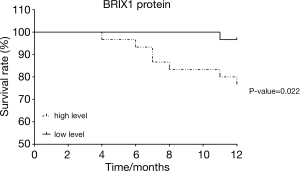
Table 2
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Elevated BRIX1 | 8.231 | 1.034–58.763 | 0.041 | 4.988 | 1.018–21.119 | 0.048 | |
| Age (years) | 1.029 | 0.968–1.094 | 0.357 | – | – | – | |
| Sex | 0.598 | 0.150–2.393 | 0.468 | – | – | – | |
| Rectal cancer | 1.195 | 0.531–2.691 | 0.667 | – | – | – | |
| Tumor stage III or IV | 9.517 | 1.170–77.402 | 0.035 | 6.711 | 1.097–36.184 | 0.044 | |
| Poor differentiation | 7.138 | 1.700–29.977 | 0.007 | 5.423 | 1.272–23.124 | 0.022 | |
| Elevated CEA | 0.766 | 0.183–3.206 | 0.715 | – | – | – | |
| Elevated CA199 | 0.615 | 0.147–2.575 | 0.506 | – | – | – | |
BRIX1, biogenesis of ribosomes BRX1; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; CI, confidence interval; HR, hazard ratio.
Discussion
The BRIX1 gene has been known to be involved in the synthesis of ribosomal 60S subunits. However, its function in the progression of cancer, especially CRC, has not been investigated. The aim of the present study was to determine the relationship between the expression of BRIX1 and the progression and prognosis of CRC patients. The results indicated that CRC tissues expressed more BRIX1 mRNA and protein than normal tissues. Moreover, the expressions of BRIX1 mRNA and protein were much higher in CRC tissues from patients with stages III–IV CRC than stages I–II CRC. An elevated level of the BRIX1 protein also acted as a risk factor of mortality in CRC patients. In the present study, we preliminarily discussed the role of BRIX1 in CRC and laid a foundation for future research of new therapeutic targets for CRC.
As an energy-intensive process in normal cells, protein synthesis is strictly regulated (14). Ribosome is essentially a supramolecular complex of RNA and protein, thus ribosome synthesis is also a part of protein synthesis (15). Four steps are needed for protein synthesis––initiation, elongation, termination, and ribosome recycling––and the step of initiation is the key step for protein synthesis. BRIX1 is involved in the synthesis of ribosomal 60S subunits, which is required at the last stage of initiation to assemble an elongation-competent 80S ribosome (16). To some degree, BRIX1 plays an important role as a key regulatory factor in protein and ribosome synthesis. However, ribosome synthesis is usually abnormal in cancer cells. Deregulated ribosome biogenesis was first found in CRC in the 1980s; genes encoding ribosome-related proteins were overexpressed in CRC cell lines and human tissues (17,18). Since then, many studies have pointed out the abnormal expression of several genes encoding ribosome-related proteins in CRC (19,20). The upregulated expression of ribosome-related proteins promotes the development, migration, and proliferation of CRC cells (20). In their study, Wang et al. reported that the knockdown of RPS24, which is responsible for the synthesis of a ribosome-related protein, inhibits the migration and proliferation of CRC cell in vitro (21). The increase of ribosome-related proteins can further induce increased ribosome synthesis. They found that, similar to our study, the expression of BRIX1 increased in CRC, which meant an upregulation of ribosome synthesis. According to the results from the Cox proportional hazards regression analysis, the upregulation of ribosome synthesis accelerated the development of CRC and increased patient mortality during follow-up.
Some commonly used chemotherapeutics for CRC, such as 5-fluorouracil (5-FU) and oxaliplatin, have been reported to interfere with not only DNA synthesis but also ribosome synthesis (22,23). 5-FU is able to combine with all kinds of RNA to alter the metabolism of RNA and reduce the biogenesis of ribosome (22). Oxaliplatin is also able to capture the components related to ribosome synthesis in CRC cells (23). The interference of these drugs on ribosome synthesis may also play a role in the treatment of CRC. However, this warrants further investigation.
As previously reported, several gene mutations in CRC may contribute to the upregulation of ribosome synthesis. The most common mutation related to ribosome synthesis in CRC is the MYC mutation. There are three kinds of RNA polymerases involved in ribosome synthesis through the transcription of rRNA. The MYC mutation was found to induce the transcriptions of all three RNA polymerases and further stimulate rRNA synthesis (24,25). Another common mutation in CRC is found in KRAS. The expression of KRAS leads to the activation of the mitogen-activated protein kinase signaling pathway, and further participates in the regulation of ribosome synthesis. Moreover, KRAS mutation was found to increase the expression of ribosome-related proteins and the translation of mRNA (26). Drugs have been developed to target these two mutations. CX-3543 and CX-5461, two specific RNA polymerase I inhibitors, have proved to be effective in some hematologic and solid malignancies in I/II clinical trials (9). They can also inhibit the proliferation of CRC, which has defects in DNA damage repair (27). KRAS mutation was once considered untreatable with drugs. Fortunately, Canon et al. developed the first KRAS inhibitor, AMG 510, in 2019 (28). It was reported to cure mice with isogenic KRAS tumors, and demonstrated anti-tumor activity in the first dosing cohort in clinical trials (28). However, the effects of AMG 510 on ribosome synthesis are unclear. According to the results of the present study, BRIX1 may be a novel target for the treatment of CRC.
The present study had some limitations. First, we used a relatively small sample size, so further analysis of a deeper relationship between BRIX1 and CRC would be difficult. Second, immunohistochemistry was not performed in the present study. Immunohistochemistry may be beneficial to determine the location of the BRIX1 protein aggregation in cancer cells and to improve our knowledge of the mechanism of how BRIX1 affects the progression and development of CRC. Third, some parameters of surgical treatment may also affect the prognosis of patients, and this will need to be improved in future research.
Conclusions
The results of the present study indicate that BRIX1 expression was elevated in CRC tissues than in adjacent normal tissues. The elevated expression of BRIX1 was also positively correlated with CRC tumor stage and contributed to a lower overall survival rate for CRC patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2564
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2564
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2564). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Affiliated Nanjing Jiangbei Hospital of Nantong University (No. J2017034) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Gbolahan O, O’Neil B. Update on systemic therapy for colorectal cancer: biologics take sides. Transl Gastroenterol Hepatol 2019;4:9. [Crossref] [PubMed]
- Liu F, Zhao J, Li C, et al. The unique prognostic characteristics of tumor deposits in colorectal cancer patients. Ann Transl Med 2019;7:769. [Crossref] [PubMed]
- Tsai HL, Huang CW, Ma CJ, et al. An observational study of vascular endothelial growth factor inhibitors as second-line treatment for metastatic colorectal cancer treated with bevacizumab plus FOLFIRI beyond progression: the association with RAS mutation and tumor sidedness. Transl Cancer Res 2019;8:2357-70. [Crossref]
- Ehrenberg R, Halama N. FOLFOX plus cetuximab in first-line therapy of advanced colorectal cancer. Ann Transl Med 2018;6:S96. [Crossref] [PubMed]
- Messersmith WA. NCCN Guidelines Updates: Management of Metastatic Colorectal Cancer. J Natl Compr Canc Netw 2019;17:599-601. [PubMed]
- Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015;14:130-46. [Crossref] [PubMed]
- Bustelo XR, Dosil M. Ribosome biogenesis and cancer: basic and translational challenges. Curr Opin Genet Dev 2018;48:22-9. [Crossref] [PubMed]
- Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 2018;18:51-63. [Crossref] [PubMed]
- Eisenhaber F, Wechselberger C, Kreil G. The Brix domain protein family -- a key to the ribosomal biogenesis pathway? Trends Biochem Sci 2001;26:345-7. [Crossref] [PubMed]
- Yang X, Feng L, Zhang Y, et al. Integrative analysis of methylome and transcriptome variation of identified cardiac disease-specific genes in human cardiomyocytes after PM2.5 exposure. Chemosphere 2018;212:915-26. [Crossref] [PubMed]
- Feng L, Yang X, Asweto CO, et al. Genome-wide transcriptional analysis of cardiovascular-related genes and pathways induced by PM2.5 in human myocardial cells. Environ Sci Pollut Res Int 2017;24:11683-93. [Crossref] [PubMed]
- Jung HM, Choi SJ, Kim JK. Expression profiles of SV40-immortalization-associated genes upregulated in various human cancers. J Cell Biochem 2009;106:703-13. [Crossref] [PubMed]
- Kafri M, Metzl-Raz E, Jona G, et al. The Cost of Protein Production. Cell Rep 2016;14:22-31. [Crossref] [PubMed]
- Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol 2009;21:435-43. [Crossref] [PubMed]
- Unbehaun A, Borukhov SI, Hellen CU, et al. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 2004;18:3078-93. [Crossref] [PubMed]
- Chester KA, Robson L, Begent RH, et al. Identification of a human ribosomal protein mRNA with increased expression in colorectal tumours. Biochim Biophys Acta 1989;1009:297-300. [Crossref] [PubMed]
- Pogue-Geile K, Geiser JR, Shu M, et al. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol 1991;11:3842-9. [Crossref] [PubMed]
- Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol 2016;17:236. [Crossref] [PubMed]
- Dong Z, Jiang H, Liang S, et al. Ribosomal Protein L15 is involved in Colon Carcinogenesis. Int J Med Sci 2019;16:1132-41. [Crossref] [PubMed]
- Wang Y, Sui J, Li X, et al. RPS24 knockdown inhibits colorectal cancer cell migration and proliferation in vitro. Gene 2015;571:286-91. [Crossref] [PubMed]
- Bash-Imam Z, Thérizols G, Vincent A, et al. Translational reprogramming of colorectal cancer cells induced by 5-fluorouracil through a miRNA-dependent mechanism. Oncotarget 2017;8:46219-33. [Crossref] [PubMed]
- Bruno PM, Liu Y, Park GY, et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med 2017;23:461-71. [Crossref] [PubMed]
- Gaviraghi M, Vivori C, Tonon G. How Cancer Exploits Ribosomal RNA Biogenesis: A Journey beyond the Boundaries of rRNA Transcription. Cells 2019;8:1098. [Crossref] [PubMed]
- Campbell KJ, White RJ. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med 2014;4:a018408. [Crossref] [PubMed]
- Charitou T, Srihari S, Lynn MA, et al. Transcriptional and metabolic rewiring of colorectal cancer cells expressing the oncogenic KRAS(G13D) mutation. Br J Cancer 2019;121:37-50. [Crossref] [PubMed]
- Reilly NM, Novara L, Di Nicolantonio F, et al. Exploiting DNA repair defects in colorectal cancer. Mol Oncol 2019;13:681-700. [Crossref] [PubMed]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
(English Language Editor: R. Scott)


