
COL4A family: potential prognostic biomarkers and therapeutic targets for gastric cancer
Introduction
Gastric cancer (GC) is the fourth most common malignancy and remains the second leading cause of cancer-related deaths (1). GC is a multifactorial disease, including environmental and genetic factors (2,3). Despite considerable advancements in prevention, diagnosis and treatment, the disease is still a great threat to human health with a five-year overall survival (OS) rate of less than 30% (4,5). Therefore, potential targets for treatments and new biomarkers for the prognosis of GC should be identified.
Conventional biomarkers (CEA, CA19-9, AFP and CA125) have been applied in diagnosis and prediction of prognosis for GC in clinical practice. Then the first molecular biomarker, HER2, is also available to improve recurrence and the efficacy of treatments. The fibroblast growth factor receptor 2 (FGFR2), vascular endothelial growth factor, E-cadherin, and TP53, etc. are recognized as metastasis related genes and could be biomarkers for recurrence forecast and metastasis assessment in GC patients (6). Besides, Immune checkpoint receptors ligands (PD-L1/2) and microsatellite-high (MSI-High) may serve as prognostic biomarkers for treatment response for GC (7). Recently, with the progression of liquid biopsy, more and more researches focus on using body fluids to detect GC biomarkers. Circulating tumor cells (CTCs), circulating cell-free DNA (cfDNA) such as EBV DNA, microRNAs such as miR-21 and miR-23a, long noncoding RNAs such as ncRuPAR, GACAT1, and GACAT2, and exosomes may provide prognostic and predictive markers for GC (8,9). With the development of knowledge of novel approach, such as TCGA Research Network, high specific and sensitive markers will continue to be tested.
The basement membrane (BM) acts as a physical barrier for prohibiting invasion and metastasis of tumors. The type IV collagen alpha chain (COL4A) family is a major component of BM that may be involved in tumor angiogenesis and progression. COL4A family constitutes of six genetically different alpha chains, α1(IV) to α6(IV), also known as COL4A1 to COL4A6 (10). COL4A1 and COL4A2 are ubiquitous, whereas COL4A3 to COL4A6 are tissue-specific. Six COL4A proteins have been found in mammalian cells, numbered according to the abundance and tissue distribution (11). COL4A1 and COL4A2 are major types and COL4A3 to COL4A6 are minor types. Mutations of COL4As genes have been confirmed to result in defective BM synthesis diseases like Goodpasture Syndrome, Alport Syndrome, and thin BM nephropathy (12-14). However, abnormal expressions of COL4As proteins have been reported involved in not only proliferation and malignant transformation but also migration and invasion of cancers by several studies (15-20).
It has been observed that up-regulated COL4A1 was closely associated with tumor growth and metastasis in papillary thyroid carcinoma (16) and promoted the proliferation of the invasive ductal carcinoma cells in breast (15). Inhibition of miR-29c could upregulation the expression of COL4A1 and increase proliferation of endometrial cancer cells (21). The suppression of COL4A2 could also significantly inhibit the migration and proliferation of triple-negative breast cancer cells (19). Compared with patients with extrahepatic bile duct carcinoma of positive COL4A2 and COL4A6, loss of COL4A2 and COL4A6 had significantly poorer prognosis (22). Nie et al. (20) reported that aberrant expression of COL4A3 might play a role in the malignant transformation of gastric epithelial cells, which is a key step in the progression of gastric carcinogenesis. COL4A4 has been observed to be downregulated in esophageal cancer (17). COL4A5 may promote lung cancer progression through discoidin domain receptor-1 (23). Ikeda et al. (18) showed that COL4A5 and COL4A6 were under-expressed in colorectal cancer as compared to normal colorectal tissues and that might remodel the epithelial BM during cancer cell invasion. Baba et al. (24) demonstrated that the expressions of COL4A5 and COL4A6 were closely related to the grade of histological atypia and tumor cell growth activity in gastric intramucosal carcinoma.
Thus, it can be inferred that COL4As family are closely related to the progression of many kinds of cancers, including gastrointestinal cancers. COL4A2 and COL4A6 may be prognostic biomarkers in extrahepatic bile duct carcinoma whereas COL4A5 and COL4A6 may be prognostic biomarkers in gastric intramucosal carcinoma. Although COL4As family are considered to be GC-related factors (20,25), the underlying mechanisms by which the COL4A factors are activated or suppressed, and their separate function in GC have not been elucidated so far.
In this study, the relationship between COL4A factors and GC was further explored. With the development of microarray technology, RNA and DNA research has been revolutionized as an essential method of biological and biomedical studies (26). The expression levels and alterations of different COL4A factors in GC patients were analyzed to identify their expression patterns, the potential functions and distinct prognostic values in GC based on the thousands of gene expression or copy number variation analysis published online.
Methods
ONCOMINE analysis
ONCOMINE gene expression array datasets (www.oncomine.org), an online cancer microarray database, was used to analyze the mRNA expression levels of COL4As in different cancers. The mRNA expressions of COL4As in clinical cancer specimens were compared with that in normal controls, using a Students’ t-test to generate a P value. The cut-off of P value and fold change was defined as 1E-4 and 2, respectively.
Gene Expression Profiling Interactive Analysis (GEPIA) dataset
GEPIA (http://gepia.cancer-pku.cn/) is a developed interactive web server for analyzing the mRNA expression data. It consists of 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline. GEPIA provides customizable functions such as tumor/normal differential expression analysis, profiling according to the cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis (27).
XENA analysis
The clinical and pathological data of 380 cases of GC patients and 37 adjacent normal tissues as well as the relative expression levels of COL4As were downloaded from TCGA 2015 RNA sequencing database from UCSC Xena (http://xena.ucsc.edu/). The mRNA expressions of COL4As in clinical cancer specimens were compared with that in normal controls. The clinical and pathological data were compared between high expression of COL4As in GC patients and low expression of that in GC patients. Statistical analyses were carried out by using SPSS 22.0 (IBM, SPSS, Chicago, IL, USA) and GraphPad Prism 7. Student’s t-test or Chi-square test was used to assess the statistical significance for comparisons of two groups (28). P value <0.05 was considered as significant.
OncoLnc tool
OncoLnc (http://www.oncolnc.org) is a tool that contains survival data for 8,647 patients from 21 cancer studies performed by The Cancer Genome Atlas (TCGA), along with RNA-SEQ expression for mRNAs and miRNAs from TCGA, and lncRNA expression from MiTranscriptome beta. It can be used to interactively explore survival correlations and to download clinical data coupled to expression data for mRNAs, miRNAs, or lncRNA. Users can investigate the range of expression of the gene at the Kaplan-Meier plotting page (29).
TCGA and CBioPortal analysis
The frequency of the COL4A family gene alterations (amplification, deep deletion, and missense mutations) and copy number variance were obtained from the cBioPortal (http://www.cbioportal.org/) (30). Besides, according to the online instructions of cBioPortal, we performed co-expression and network analyses.
Functional enrichment analysis
In this study, Metascape (http://metascape.org) was used to conduct pathway and process enrichment analysis of COL4A family members and neighboring genes significantly associated with COL4As alterations (31). The Gene Ontology (GO) terms as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched based on the Metascape online tool. PPI enrichment analysis was performed. Further, MCODE (Molecular Complex Detection) algorithm was applied to identify densely connected network components.
Results
The mRNA expression levels of COL4As in patients with GC
ONCOMINE database was used in this study to compare the mRNA expression levels of COL4As in cancers to those in normal tissues (Figure 1). It was found that the mRNA expression level of COL4A1 was significantly upregulated in GC patients in six datasets. In Chen’s dataset (32), the expression level of COL4A1 was significantly up-regulated in all types of GC as compared to that in normal tissues (in diffuse gastric adenocarcinoma with a fold change of 5.045, gastric intestinal-type adenocarcinoma of 4.104, and gastric mixed adenocarcinoma of 6.23, Table 1). COL4A2 was also overexpressed with a fold change of 10.501, 2.113, and 3.82 in diffuse gastric adenocarcinoma, gastric intestinal-type adenocarcinoma, and gastric mixed adenocarcinoma respectively in Chen’s dataset as shown in Table 1. D-Errico (33) showed another increased expression of factor, COL4A4 in diffuse gastric adenocarcinoma with a fold change of 2.739 compared with normal samples (Table 1).
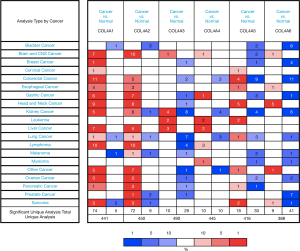
Table 1
| Types of GC vs. gastric | Fold change | P value | t-test | Ref | |
|---|---|---|---|---|---|
| COL4A1 | Diffuse gastric adenocarcinoma vs. normal | 5.045 | 4.54E-13 | 14.254 | Chen |
| Gastric mixed adenocarcinoma vs. normal | 6.23 | 6.43E-07 | 10.438 | Chen | |
| Gastric intestinal type adenocarcinoma vs. normal | 4.104 | 6.04E-18 | 15.779 | Chen | |
| Total gastric cancer vs. normal | 2.276 | 5.67E-06 | 5.853 | Wang | |
| COL4A2 | Diffuse gastric adenocarcinoma vs. normal | 10.501 | 1.63E-09 | 10.501 | Chen |
| Gastric intestinal type adenocarcinoma vs. normal | 2.133 | 2.38E-17 | 10.669 | Chen | |
| Gastric mixed adenocarcinoma vs. normal | 3.82 | 4.23E-06 | 9.131 | Chen | |
| Total gastric cancer vs. normal | 2.483 | 1.85E-06 | 5.93 | Wang | |
| COL4A3 | Gastric intestinal type adenocarcinoma vs. normal | –2.526 | 1.03E-05 | –4.775 | DErrico |
| COL4A4 | Diffuse gastric adenocarcinoma vs. normal | 2.739 | 1.26E-05 | 5.168 | DErrico |
| COL4A5 | Diffuse gastric adenocarcinoma vs. normal | –2.585 | 9.71E-08 | –6.25 | Cho |
| Gastric intestinal type adenocarcinoma vs. normal | –4.467 | 7.41E-05 | –4.467 | Cho | |
| COL4A6 | Gastric intestinal type adenocarcinoma vs. normal | –2.748 | 2.26E-08 | –6.522 | Derrico |
On the contrary, in the same dataset of D-Errico, the expression levels of COL4A3 and COL4A6 were significantly decreased in gastric intestinal-type adenocarcinoma with a fold change of −2.526 and −2.748 respectively (Table 1). As shown in Table 1 for COL4A5, there was down-regulation of mRNA expressions in diffuse gastric adenocarcinoma and gastric intestinal-type adenocarcinoma in comparison with normal patients with −2.585 and −4.467 fold changes separately according to Cho’s dataset (34).
The relationship between mRNA levels of COL4As and clinicopathological parameters of GC patients
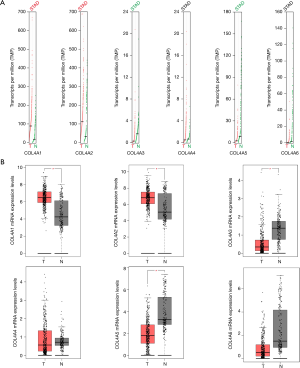
By using GEPIA dataset, a comparison of the mRNA expression of COL4A factors between GC and gastric tissues. It was indicated from the results that COL4A1 and COL4A2 expressions were up-regulated in gastric adenocarcinoma compared with gastric tissues, whereas the expression levels of COL4A3 and COL4A5 were lower in gastric adenocarcinoma than gastric tissues (Figure 2).
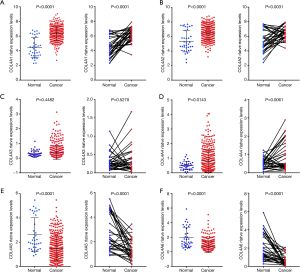
In order to further validate the relationship between mRNA levels of COL4As and clinicopathological parameters of GC patients, we downloaded the clinicopathological data of 380 cases of GC patients and 37 adjacent normal tissues as well as the relative mRNA expression levels of COL4As from UCSC Xena database (28). The clinicopathological data of 380 cases of GC patients is shown in Table S1. By using TCGA sequencing data, we further validated that there was the increased mRNA expressions of COL4A1, COL4A2 and COL4A4 and the decreased mRNA expressions of COL4A5 and COL4A6 in human gastric adenocarcinoma tissues (n=380), adjacent normal tissues (n=37), and in paired GC tissues (n=34) (Figure 3).
To evaluate whether the mRNA expression levels of COL4As were associated with clinical and pathological characteristics and prognosis of GC patients, 380 GC patients were divided into two groups based on the mean of mRNA expressions of each one of COL4As. There were COL4As high expression (the value > the median) and COL4As low expression (the value ≤ the median). As indicated in Table 2, COL4A2 expression positively correlated with classification of the grade (P=0.044). High expressions of both COL4A3 and COL4A4 were linked to poor TNM stage, pathological grade, lymph node metastasis, and Lauren’s classification (P<0.05). The expression of COL4A5 was high in patients aged less than 60 years. COL4A6 harbored no association with clinical parameters (P>0.05). Multivariate analysis using the Cox proportional hazards model indicated that age, gender, pathological grade, metastasis and COL4A5 expression are independent prognostic factors for OS. However, TNM stage, lymph node metastasis, Lauren’s classification, COL4A1-4 and COL4A6 were associated with poor OS but not independent prognostic factors (Figure S1).
Table 2
| Clinicopathologic features [cases of data available*] | Cases (n) | COL4A1 | COL4A2 | COL4A3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P | Low | High | P | Low | High | P | ||||
| Age [375] | ||||||||||||
| ≥60 | 258 | 127 | 131 | 132 | 126 | 135 | 123 | 0.181 | ||||
| <60 | 117 | 62 | 55 | 0.506 | 57 | 60 | 0.738 | 52 | 65 | |||
| Gender [380] | ||||||||||||
| Female | 131 | 67 | 64 | 68 | 63 | 68 | 63 | 0.666 | ||||
| Male | 249 | 123 | 126 | 0.829 | 122 | 127 | 0.666 | 122 | 127 | |||
| TNM stage [376] | ||||||||||||
| I | 53 | 30 | 23 | 33 | 20 | 37 | 16 | 0.003 | ||||
| II–IV | 323 | 159 | 164 | 0.374 | 155 | 168 | 0.075 | 151 | 172 | |||
| Pathological grade [371] | ||||||||||||
| G1–2 | 148 | 76 | 69 | 85 | 63 | 88 | 60 | 0.003 | ||||
| G3 | 223 | 110 | 113 | 0.460 | 104 | 119 | 0.044 | 97 | 126 | |||
| Lymph node metastasis [370] | ||||||||||||
| N0 | 117 | 57 | 60 | 64 | 53 | 72 | 45 | 0.004 | ||||
| N1–N3 | 253 | 129 | 124 | 0.738 | 122 | 131 | 0.265 | 113 | 140 | |||
| Metastasis [361] | ||||||||||||
| M0 | 341 | 173 | 168 | 174 | 167 | 173 | 168 | 1 | ||||
| M1 | 20 | 10 | 10 | 1.000 | 8 | 12 | 0.366 | 10 | 10 | |||
| Lauren classification [241] | ||||||||||||
| Intestinal type | 165 | 83 | 82 | 83 | 82 | 92 | 73 | 0.004 | ||||
| Diffuse type | 76 | 32 | 44 | 0.268 | 31 | 45 | 0.211 | 27 | 49 | |||
*, there are some data missed in clinicopathological data of GC patients from TCGA database.
To determine the COL4As protein expressions, they were analyzed using clinical specimens retrieved from the Human Protein Atlas (the Human Protein Atlas available from www.proteinatlas.org). It showed that COL4A1 and COL4A2 had strong expressions in GC and weak expressions in normal tissues (Figure S2). Whereas COL4A3 had the inverse expression (Figure S2). Unfortunately, related results of other COL4As have not been uploaded till date; hence, they could not be presented here.
Increased mRNA expressions of COL4A1/2/5 were associated with OS of GC patients and increased mRNA expressions of COL4A3/4 were associated with poor disease-free survival (DFS) of GC patients (Figure 4)
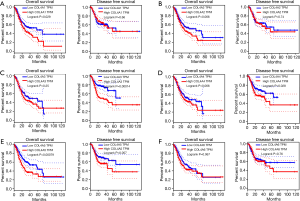
COL4A1 and COL4A5 high mRNA expression levels were found to be associated with poor OS of patients in GC by using both GEPIA and OncoLnc tools (29) (Figure 4A,E, Figure S3). The high mRNA expression of COL4A2 was correlated with poor OS of patients in GC by using OncoLnc. However, results not same with GEPIA tool (Figure 4B, Figure S3). Different results between GEPIA and OncoLnc tools may derived from different cut-off values. The high expressions of COL4A3 and COL4A4 were found to be associated with poor DFS of patients in GC (Figure 4C,D). Other factors had no links with OS or DFS of patients in GC (Figure 4F).
Predicted functions and pathways of the changes in COL4A factors and their frequently altered neighboring genes in patients with GC
The cBioPortal online tool (30,35) was used to analyze the COL4As alterations, correlations, and networks for GC (TCGA, Provisional). There were 152 samples out of 478 patients of COL4As altering with GC (32%). In almost half of the samples (72 samples), two or more alterations were detected (Figure 5A). The percentages of genetic alterations in individual genes of COL4A family members for GC varied from 6% to 13% (COL4A1, 13%; COL4A2, 11%; COL4A3, 7%; COL4A4, 7%; COL4A5, 8%; COL4A6, 6%; Figure 5A). The correlations of COL4As with each other were calculated by analyzing their mRNA expressions (RNA Seq V2 RSEM) via the online tool mentioned above. Pearson’s correction was included. The results indicated significant and positive correlations in the following COL4As: COL4A1 with COL4A2, COL4A3 with COL4A4, and COL4A5 with COL4A6 (Figure 5B). Then we performed network analysis for COL4As and the 50 most frequently altered neighboring genes (Figure 5C). The names, abbreviations, and functions for these genes are shown in Table S2. The results showed that the integrin family of protein-coding genes (ITGA2B/3/4/6/7/E/L/V/X, ITGB4/5/7/8/L1) and the laminin family of protein-coding genes (LAM1/2/3/4/5, LAMB1/2/3/4, LAMC1/3) were closely associated with COL4As alterations.
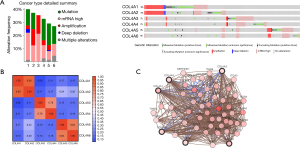
Functional gene and pathway enrichment analysis of COL4A factors and their frequently altered neighboring genes in patients with GC
The GO functions and KEGG pathway enrichment analysis of candidate COL4As and their frequently altered neighboring genes was performed based on the Metascape databases (31). As shown in Figure 6A, there were the top 20 GO and KEGG enrichment items (17 terms and 3 pathways) involved. It was indicated that these COL4As and their frequently altered neighboring genes were mainly enriched in extracellular matrix organization, integrin-mediated signaling pathway, endoplasmic reticulum lumen, endodermal cell differentiation, cell morphogenesis involved in differentiation, cell junction assembly, positive regulation of cell migration, blood vessel morphogenesis, platelet degranulation, laminin-5 complex, and laminin-1 complex, etc. Three significantly enriched pathways: focal adhesion, toxoplasmosis, and proteoglycans in cancer were identified in correlations with COL4As and their frequently altered neighboring genes. Furthermore, these enriched terms were closely connected with each other and clustered into intact networks (Figure 6B).
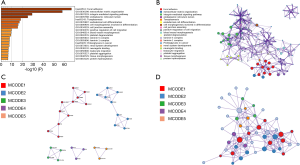
For a better understanding, the relationship between COL4A family members and GC, the Metascape database was used to perform protein-protein interaction (PPI) enrichment analysis. The PPI network is shown in Figure 6C,D. The five most significant MCODE components were extracted from the PPI network. Each MCODE component was applied by pathway and process enrichment analysis independently, and the three best-scoring terms of the corresponding components by P value were retained as the functional description shown in Table 3. The results suggested that ECM-receptor interaction, PID integrin pathway, extracellular matrix organization, focal adhesion, laminin interactions, and cell junction assembly were mainly associated with COL4A family members.
Table 3
| MCODE | GO | Description | Log10(P) |
|---|---|---|---|
| MCODE_1 | hsa04512 | ECM-receptor interaction | –23.9 |
| MCODE_1 | M18 | PID INTEGRIN1 PATHWAY | –21.6 |
| MCODE_1 | R-HSA-1474244 | Extracellular matrix organization | –21 |
| MCODE_2 | hsa04510 | Focal adhesion | –18.8 |
| MCODE_2 | R-HSA-3000157 | Laminin interactions | –12.6 |
| MCODE_2 | GO:0034329 | Cell junction assembly | –12.5 |
| MCODE_3 | R-HSA-186797 | Signaling by PDGF | –10.5 |
| MCODE_3 | R-HSA-3000171 | Non-integrin membrane-ECM interactions | –10.5 |
| MCODE_3 | R-HSA-2214320 | Anchoring fibril formation | –9.1 |
| MCODE_4 | CORUM:6990 | THSD1-FAK-talin-vinculin complex | –11.8 |
| MCODE_4 | CORUM:5177 | Polycystin-1 multiprotein complex (ACTN1, CDH1, SRC, JUP, VCL, CTNNB1, PXN, BCAR1, PKD1, PTK2, TLN1) | –10.2 |
| MCODE_4 | M281 | PID FAK PATHWAY | –7.9 |
| MCODE_5 | R-HSA-1650814 | Collagen biosynthesis and modifying enzymes | –7.7 |
| MCODE_5 | R-HSA-1474290 | Collagen formation | –7.3 |
| MCODE_5 | R-HSA-1474244 | Extracellular matrix organization | –5.7 |
Discussion
In this study, the mRNA expression levels, prognostic values, genetic alterations, correlations, and potential functions of different COL4As in GC, were systematically explored by bioinformatics analysis.
COL4A1, the most classic member of the COL4A family is found to play a pivotal role in proliferation, metastasis, and invasion in most cancers (15,16,36). Zhang et al. (37) reported that miRNA-29c-3p represses proliferation of gastric adenocarcinoma BGC-823 cells by directly targeting COL4A1. Huang et al. (38) identified that COL4A1 is upregulated in trastuzumab resistance in GC cells and may induce trastuzumab resistant in GC in silico. In the present study, it was revealed that the mRNA expression of COL4A1 was upregulated in human GC as compared to normal tissues. High mRNA expression level of COL4A1 was found to be associated with poor OS by using both GEPIA and OncoLnc tool. However, expression of COL4A1 did not correlate with the DFS and clinical characteristics of the patients with GC. These phenomena indicate COL4A1 may serve as a new biomarker for the prognosis and a potential target of GC. As demonstrated in Figure 1, COL4A1 is also upregulated in lymphoma and sarcoma. Moreover, Chida et al. (39) identified that COL4A1 is located predominantly in cancer stroma. Immunohistochemistry (Figure S2) also showed COL4A1 is expressed in stromal tissue but not in tumor cells. In addition, COL4A1 may be derived from stromal reaction during the tumor progression and not from tumor cells themselves. Miyake et al. (36) found that the formation of tumor budding is involved in the carcinogenesis of COL4A1 in human urothelial cancer of the bladder. However, in papillary thyroid cancer, exosomal miR-21-5p can increase endothelial tube formation by inhibiting COL4A1, consequently promoting angiogenesis (40). Angiogenesis may be involved in the effects of COL4A1 on tumors. Additional experiments need be performed in the future to elucidate whether COL4A1 promotes or inhibits angiogenesis in GC.
In triple-negative breast cancer, knockdown of COL4A2 could inhibit the proliferation and migration of cancer cells (19). COL4A2 is identified as a methylation marker with high accuracy for the detection of colorectal cancer (41). Notch3 can upregulate COL4A2 and promote anoikis resistance in ovarian cancer (42). In our study, use of ONCOMINE, GEPIA, and UCSC Xena datasets also revealed the mRNA expression level of COL4A2 was up-regulated in GC. By using the GEPIA tool, it was analyzed that COL4A2 was not related to OS and DFS of patients in GC. High transcriptional expression level of COL4A2 was associated with poor OS in OncoLnc. Moreover, COL4A2 high expression positively correlated with pathological grade (P=0.044). These findings suggested that COL4A2 may be a promising therapeutic target and could predict the prognosis of patients in GC as a biomarker.
Metodieva et al. (43) found that COL4A3 is downregulated in early-stage non-small cell lung cancer by real-time PCR. COL4A3 expressed to a lesser extent in GC than in normal mucosa at both mRNA and protein levels but its expression positively related to poor prognosis and worse clinicopathologic features of GC (20). A decreased mRNA expression of COL4A3 in GC is also shown in our study. Both GEPIA and OncoLnc results showed COL4A3 was not related to OS of patients in GC. However, to our surprise, high COL4A3 mRNA expression was significantly associated with poor DFS and the high mRNA expressions of COL4A3 was correlated with poor TNM stage, pathological grade, lymph node metastasis, and Lauren’s classification (P<0.05). It seemed COL4A3 might serve as a biomarker to indicate a worse prognosis of patients in GC.
COL4A4 was confirmed to be down-regulated in esophageal cancer (17). However, in the present study, excluding GEPIA datasets, ONCOMINE and UCSC Xena showed an increased mRNA expression of COL4A4 in GC. High COL4A4 mRNA expression was associated with poor DFS and poor TNM stage, pathological grade, lymph node metastasis and Lauren classification (P<0.05). There was no association between COL4A4 expression level and OS of patients in GC. COL4A4 may play a role of an oncogene and a potential prognostic biomarker for GC.
ONCOMINE, GEPIA and UCSC Xena datasets all revealed that the mRNA expression of COL4A5 was downregulated in GC. Except for GEPIA dataset, ONCOMINE, and XENA datasets showed low mRNA expression of COL4A6 in GC compared with normal samples. COL4A5 was not related to DFS. We found high mRNA expression of COL4A5 was correlated with poor OS. Multivariate analysis indicated that age, gender, pathological grade, metastasis, and COL4A5 expression are independent prognostic factors. COL4A6 harbored no link with OS, DFS and clinical characteristics. Loss of expressions of COL4A5 and COL4A6 were reported in colorectal cancer and might be involved in the remodeling of the epithelial BM during cancer cell invasion (18). COL4A5 may be an indicator of a worse prognosis of GC. Further research is needed to prove whether COL4A6 plays a role in GC.
The percentages of genetic alterations in COL4A family members for GC were calculated to further illustrate the genetic alterations, potential functions, and carcinogenic mechanisms of the same. The percentages of genetic alterations ranged from 6% to 13% for individual genes based on TCGA Provisional dataset. In addition, we predicted COL4As alterations related 50 genes and constructed a network. COL4As alterations were closely associated with the integrin family of protein-coding genes, including ITGA2B/3/4/6/7/E/L/V/X, ITGB4/5/7/8/L1, and the laminin family of protein-coding genes, including LAM1/2/3/4/5, LAMB1/2/3/4, LAMC1/3. The GO and KEGG pathway analysis indicated they were enriched in pathways between ECM and the adhesion process. Adhesion-related pathways, such as extracellular matrix organization, focal adhesion and integrin-mediated signaling pathways were associated with the processes of proliferation, migration and invasion of GC (44-46). Combined with the results above, we hypothesized that COL4As may have impacts on adhesion-related pathways and integrin-mediated signaling pathways, thereby regulating the downstream of Akt pathway. Activation of Akt pathway could promote the proliferation and invasion of GC.
This study is a descriptive research using bioinformatics analysis. In future, a large sample sizes with high quality and experimental studies in our hospital are needed to further elucidate and verify our research.
Conclusions
In this study, we used the GEPIA tool, cBioPortal, and Metascape tool to explore the expression and prognostic value of COL4As in GC from which we could have a further understanding of the molecular biological properties of GC. Our findings suggested that COL4A1/2 are potential therapeutic targets for GC, COL4A3/4/6 may have an impact on gastric carcinogenesis and subsequent progression and COL4A5 is found to be an independent prognostic marker for GC.
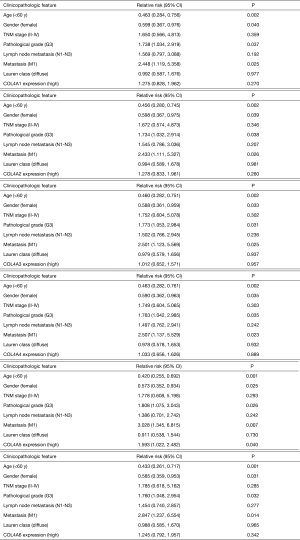

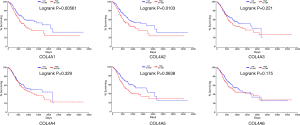
Table S1
| Parameters [cases of data available*] | Cases (n=380) |
|---|---|
| Age [375] | |
| ≥60 | 258 (68.8) |
| <60 | 117 (31.2) |
| Gender [380] | |
| Female | 131 (34.5) |
| Male | 249 (65.5) |
| Pathological stage [368] | |
| I/II | 171 (46.5) |
| III/IV | 197 (53.5) |
| T classification [377] | |
| T1/T2 | 98 (26.0) |
| T3/T4 | 279 (74.0) |
| N classification [370] | |
| N0/N1 | 218 (58.9) |
| N2/N3 | 152 (41.1) |
| Metastasis and[or] recurrence [380] | |
| Negative | 315 (82.9) |
| Positive | 65 (17.1) |
*, there are some data missed in clinicopathological data of GC patients from TCGA database. Data present as n (%).
Table S2
| NO. | Gene symbol | Full name | Function |
|---|---|---|---|
| 1 | ACTB | Actin beta | Cell motility, structure, integrity, and intercellular signaling |
| 2 | ACTN2 | Actinin alpha 2 | Bind actin to the membrane, anchor the myofibrillar actin filaments |
| 3 | ACTN4 | Actinin alpha 4 | Bind actin to the membrane, anchor the myofibrillar actin filaments |
| 4 | CASP8 | Caspase 8 | Be involved in the programmed cell death induced by Fas and various apoptotic stimuli |
| 5 | COL18A1 | Collagen type XVIII alpha 1 chain | Inhibit angiogenesis and tumor growth |
| 6 | COL21A1 | Collagen type XXI alpha 1 chain | Maintain the integrity of the extracellular matrix |
| 7 | COL22A1 | Collagen type XXII alpha 1 chain | Contribute to the stabilization of myotendinous junctions and strengthen skeletal muscle attachments during contractile activity |
| 8 | COL26A1 | Collagen type XXVI alpha 1 chain | Be associated with aspirin-intolerant asthma |
| 9 | COL28A1 | Collagen type XXVIII alpha 1 chain | Belong to a class of collagens containing von Willebrand factor type A (VWFA) domains |
| 10 | COL7A1 | Collagen type VII alpha 1 chain | Function as an anchoring fibril between the external epithelia and the underlying stroma |
| 11 | FLNA | Filamin A | Be involved in remodeling the cytoskeleton to effect changes in cell shape and migration; interact with integrins, transmembrane receptor complexes, and second messengers |
| 12 | FLNB | Filamin B | Repair vascular injuries |
| 13 | FN1 | Fibronectin 1 | Be involved in cell adhesion and migration processes including embryogenesis, wound healing, blood coagulation, host defense, and metastasis |
| 14 | ITGA2B | Integrin subunit alpha 2b | Blood coagulation |
| 15 | ITGA3 | Integrin subunit alpha 3 | |
| Form an integrin that interacts with extracellular matrix proteins including members of the laminin family; be correlated with breast cancer metastasis | |||
| 16 | ITGA4 | Integrin subunit alpha 4 | May play a role in cell motility and migration |
| 17 | ITGA6 | Integrin subunit alpha 6 | Function in cell surface adhesion and signaling |
| 18 | ITGA7 | Integrin subunit alpha 7 | Play a role in cell migration, morphologic development, differentiation, and metastasis |
| 19 | ITGAE | Integrin subunit alpha E | Adhesion; serve as an accessory molecule for human intestinal intraepithelial lymphocytes activation |
| 20 | ITGAL | Integrin subunit alpha L | Leukocyte intercellular adhesion; function in lymphocyte costimulatory signaling |
| 21 | ITGAV | Integrin subunit alpha V | Regulate angiogenesis and cancer progression |
| 22 | ITGAX | Integrin subunit alpha X | Adherence of neutrophils and monocytes to stimulated endothelium cells, and phagocytosis of complement coated particles |
| 23 | ITGB4 | Integrin subunit beta 4 | Play a pivotal role in the biology of invasive carcinoma |
| 24 | ITGB5 | Integrin subunit beta 5 | Participate in cell adhesion as well as cell-surface mediated signaling |
| 25 | ITGB7 | Integrin subunit beta 7 | Play a role in leukocyte adhesion |
| 26 | ITGB8 | Integrin subunit beta 8 | Play a role in human airway epithelial proliferation |
| 27 | ITGBL1 | Integrin subunit beta like 1 | Contain integrin-like cysteine-rich repeats |
| 28 | LAMA1 | Laminin subunit alpha 1 | Cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis |
| 29 | LAMA2 | Laminin subunit alpha 2 | Mediate the attachment, migration, and organization of cells into tissues during embryonic development |
| 30 | LAMA3 | Laminin subunit alpha 3 | Be essential for formation and function of the basement membrane and have additional functions in regulating cell migration and mechanical signal transduction |
| 31 | LAMA4 | Laminin subunit alpha 4 | The exact function of laminin, alpha 4 is not known |
| 32 | LAMA5 | Laminin subunit alpha 5 | The major noncollagenous constituent of basement membranes |
| 33 | LAMB1 | Laminin subunit beta 1 | Inhibit metastasis |
| 34 | LAMB2 | Laminin subunit beta 2 | The maturation of neuromuscular junctions and maintain glomerular filtration |
| 35 | LAMB3 | Laminin subunit beta 3 | Belong to a family of basement membrane proteins |
| 36 | LAMB4 | Laminin subunit beta 4 | Cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis |
| 37 | LAMC1 | Laminin subunit gamma 1 | Cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis |
| 38 | LAMC3 | Laminin subunit gamma 3 | Cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis |
| 39 | MSR1 | Macrophage scavenger receptor 1 | Mediate the endocytosis of modified low-density lipoproteins; regulation of scavenger receptor activity in macrophages. |
| 40 | P3H2 | Prolyl 3-hydroxylase 2 | Collagen chain assembly, stability and cross-linking |
| 41 | P4HB | Prolyl 4-hydroxylase subunit beta | A highly abundant multifunctional enzyme |
| 42 | PDGFA | Platelet derived growth factor subunit A | Bind and activate PDGF receptor tyrosine kinases |
| 43 | PLOD2 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | Be critical for the stability of intermolecular crosslinks |
| 44 | PLOD3 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 | Be critical for the stability of intermolecular crosslinks |
| 45 | PTK2 | Protein tyrosine kinase 2 | Cell growth and intracellular signal transduction pathways |
| 46 | SERPINH1 | Serpin family H member 1 | A marker for cancer |
| 47 | THBS2 | Thrombospondin 2 | A potent inhibitor of tumor growth and angiogenesis |
| 48 | THBS4 | Thrombospondin 4 | Be activated during the stromal response to invasive breast cancer; play a role in inflammatory responses in Alzheimer's disease |
| 49 | TLN1 | Talin 1 | Assist in the attachment of adherent cells to extracellular matrices and of lymphocytes to other cells |
Acknowledgments
Funding: This study was supported by
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-517
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-517). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved. All the datasets were retrieved from the published literature and publicly available datasets. Datasets links were attached in the
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Mills JC, Samuelson LC. Past Questions and Current Understanding About Gastric Cancer. Gastroenterology 2018;155:939-44. [Crossref] [PubMed]
- Mun DG, Bhin J, Kim S, et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell 2019;35:111-24.e10. [Crossref] [PubMed]
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977-1010. [Crossref] [PubMed]
- de Mestier L, Lardiere-Deguelte S, Volet J, et al. Recent insights in the therapeutic management of patients with gastric cancer. Dig Liver Dis 2016;48:984-94. [Crossref] [PubMed]
- Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol 2018;24:2818-32. [Crossref] [PubMed]
- Jiang B, Sun Q, Tong Y, et al. An immune-related gene signature predicts prognosis of gastric cancer. Medicine (Baltimore) 2019;98:e16273. [Crossref] [PubMed]
- Li TT, Liu H, Yu J, et al. Prognostic and predictive blood biomarkers in gastric cancer and the potential application of circulating tumor cells. World J Gastroenterol 2018;24:2236-46. [Crossref] [PubMed]
- Shekari N, Baradaran B, Shanehbandi D, et al. Circulating MicroRNAs: Valuable Biomarkers for the Diagnosis and Prognosis of Gastric Cancer. Curr Med Chem 2018;25:698-714. [PubMed]
- Hudson BG, Reeders ST, Tryggvason K. Type IV collagen: structure, gene organization, and role in human diseases. Molecular basis of Goodpasture and Alport syndromes and diffuse leiomyomatosis. J Biol Chem 1993;268:26033-6. [PubMed]
- Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech 2008;71:357-70. [Crossref] [PubMed]
- Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer 2003;3:422-33. [Crossref] [PubMed]
- Gast C, Pengelly RJ, Lyon M, et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant 2016;31:961-70. [Crossref] [PubMed]
- Ozdemir G, Gulhan B, Atayar E, et al. COL4A3 mutation is an independent risk factor for poor prognosis in children with Alport syndrome. Pediatr Nephrol 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Jin R, Shen J, Zhang T, et al. The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget 2017;8:58172-83. [Crossref] [PubMed]
- Zhang H, Teng X, Liu Z, et al. Gene expression profile analyze the molecular mechanism of CXCR7 regulating papillary thyroid carcinoma growth and metastasis. J Exp Clin Cancer Res 2015;34:16. [Crossref] [PubMed]
- Chattopadhyay I, Phukan R, Singh A, et al. Molecular profiling to identify molecular mechanism in esophageal cancer with familial clustering. Oncol Rep 2009;21:1135-46. [PubMed]
- Ikeda K, Iyama K, Ishikawa N, et al. Loss of expression of type IV collagen alpha5 and alpha6 chains in colorectal cancer associated with the hypermethylation of their promoter region. Am J Pathol 2006;168:856-65. [Crossref] [PubMed]
- JingSong H. siRNA-mediated suppression of collagen type iv alpha 2 (COL4A2) mRNA inhibits triple-negative breast cancer cell proliferation and migration. Oncotarget 2017;8:2585-93. [Crossref] [PubMed]
- Nie XC, Wang JP, Zhu W, et al. COL4A3 expression correlates with pathogenesis, pathologic behaviors, and prognosis of gastric carcinomas. Hum Pathol 2013;44:77-86. [Crossref] [PubMed]
- Van Sinderen M, Griffiths M, Menkhorst E, et al. Restoration of microRNA-29c in type I endometrioid cancer reduced endometrial cancer cell growth. Oncol Lett 2019;18:2684-93. [Crossref] [PubMed]
- Hirashima K, Iyama K, Baba Y, et al. Differential expression of basement membrane type IV collagen α2 and α6 chains as a prognostic factor in patients with extrahepatic bile duct carcinoma. J Surg Oncol 2013;107:402-7. [Crossref] [PubMed]
- Xiao Q, Jiang Y, Liu Q, et al. Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1. PLoS Genet 2015;11:e1005249. [Crossref] [PubMed]
- Baba Y, Iyama K, Ikeda K, et al. Differential expression of basement membrane type IV collagen alpha chains in gastric intramucosal neoplastic lesions. J Gastroenterol 2007;42:874-80. [Crossref] [PubMed]
- Kim IW, Jang H, Kim JH, et al. Computational Drug Repositioning for Gastric Cancer using Reversal Gene Expression Profiles. Sci Rep 2019;9:2660. [Crossref] [PubMed]
- Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol 2011;671:3-34. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science 2016;2:e67. [Crossref]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell 2003;14:3208-15. [Crossref] [PubMed]
- D'Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009;45:461-9. [Crossref] [PubMed]
- Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res 2011;17:1850-7. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Miyake M, Hori S, Morizawa Y, et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017;8:36099-114. [Crossref] [PubMed]
- Zhang QN, Zhu HL, Xia MT, et al. A panel of collagen genes are associated with prognosis of patients with gastric cancer and regulated by microRNA-29c-3p: an integrated bioinformatics analysis and experimental validation. Cancer Manag Res 2019;11:4757-72. [Crossref] [PubMed]
- Huang R, Gu W, Sun B, et al. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol Med Rep 2018;17:6387-96. [Crossref] [PubMed]
- Chida S, Okayama H, Noda M, et al. Stromal VCAN expression as a potential prognostic biomarker for disease recurrence in stage II-III colon cancer. Carcinogenesis 2016;37:878-87. [Crossref] [PubMed]
- Wu F, Li F, Lin X, et al. Exosomes increased angiogenesis in papillary thyroid cancer microenvironment. Endocr Relat Cancer 2019;26:525-38. [Crossref] [PubMed]
- Liu X, Wen J, Li C, et al. High-Yield Methylation Markers for Stool-Based Detection of Colorectal Cancer. Dig Dis Sci 2020;65:1710-9. [Crossref] [PubMed]
- Brown CW, Brodsky AS, Freiman RN. Notch3 overexpression promotes anoikis resistance in epithelial ovarian cancer via upregulation of COL4A2. Mol Cancer Res 2015;13:78-85. [Crossref] [PubMed]
- Metodieva SN, Nikolova DN, Cherneva RV, et al. Expression analysis of angiogenesis-related genes in Bulgarian patients with early-stage non-small cell lung cancer. Tumori 2011;97:86-94. [Crossref] [PubMed]
- Kumar S, Kapoor A, Desai S, et al. Proteolytic and non-proteolytic regulation of collective cell invasion: tuning by ECM density and organization. Sci Rep 2016;6:19905. [Crossref] [PubMed]
- Zhou J, Yi Q, Tang L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: a focused review. J Exp Clin Cancer Res 2019;38:250. [Crossref] [PubMed]
- Yu R, Li Z, Zhang C, et al. Elevated limb-bud and heart development (LBH) expression indicates poor prognosis and promotes gastric cancer cell proliferation and invasion via upregulating Integrin/FAK/Akt pathway. PeerJ 2019;7:e6885. [Crossref] [PubMed]


