
High levels of MESP1 expression in non-small cell lung cancer can facilitate cell proliferation, metastasis and suppresses cell apoptosis
Introduction
In the United States and around the world, lung cancer (LC) causes the most cancer-related deaths in men and women, and it has become one of the most common types of cancer (1). According to the histological classification, LC is main composed of non-small cell lung cancer (NSCLC) (80%) and small cell lung cancer (SCLC). Due to several reasons of limited therapeutic choices, it has behaved malignant metastases of tumor, high recurrence rates and the overall 5-year survival rate of NSCLC is less than 15% (2). Traditional diagnosis includes small biopsy and cytological specimen tests, and treatment with bevacizumab or pemetrexed which are sensitive to lung adenocarcinoma cases without carcinogenic driver mutations. Until epidermal growth factor receptor (EGFR) mutation was found as effective targets for EGFR tyrosine inhibitors in lung adenocarcinoma patients. Studies have confirmed that AXL receptor tyrosine kinase is a potential therapeutic target, and its inhibition can prevent or overcome the acquired resistance of EGFR mutant LC patients to EGFR-TKIs(tyrosine kinase inhibitors) (3). More and more researches have focused on genetic therapy. In addition to that key gene molecules in LC, it have been reported to be associated with multiple gene families, such as receptor tyrosine kinases, epigenetic factors, transcription factors, signaling factoes, cell cycle factors and even tumor suppressors (4), which may help individualized treatment strategies for patients with advanced LC. Patients also need molecular tests to determine the type of LC and to qualify for specific therapies (5). Therefore, studying new oncogenes and finding more effective molecular biological markers are great significance for the diagnosis and treatment of NSCLC.
With the increasing application of Biochip technologies, DNA microarray analysis has become the most widely used genome-scale data source in life science research. Microarray analysis of mRNA is to compare genes in adjacent normal tissues and tumor tissues to identify genes with significant statistical differences. We performed mRNA microarray analysis on 15 pairs of NSCLC tissues and adjacent normal tissues. It was found that MESP1 had relatively high expression in the tumor tissues and showed a role in the epithelialization of somatic mesoderm and the development of cardiac mesoderm. It also participates in the cell’s positive regulation of transcription, DNA-templated process. As we known, MESP1 can activate many downstream effectors of mesoderm. By knocking down MESP1, it was found that MESP1 can identify the differentiation characteristics of early vascular progenitor cells (6). Meanwhile, MESP1 is an important transcription factor that regulates epithelial-mesenchymal transitions (EMT) and cardiovascular commitment (7), including chemotaxis and apoptosis of cells. From a phylogenetic perspective, MESP1 plays an important regulatory role in human embryonic development. Studies have shown that pathological variations of MESP1 may contribute to the occurrence of disease and participate in many physiological processes of tumors (8). However, the expression pattern and underlying molecular function of MESP1 are still elusive in NSCLC. Thus the present study was conducted to investigate the functional mechanism of MESP1 in NSCLC and whether it provides a potential clinical guideline for tumorigenesis and tumor progression. We believe the present findings will be helpful in the clinical diagnosis of LC, and provides new targets for the treatment of NSCLC patients. We present the following article in accordance with the CONSORT reporting checklist and MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1132).
Methods
Human tissue sample collection
All subjects were obtained from NSCLC patient samples collected by the Yunnan Cancer Hospital. Those above processes were approved by the patients, their families, ethics committee of the Yunnan Cancer Hospital (approval number: KY202014), and met the clinical norms and medical ethics regulations. Written informed consent was signed by every patient. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Which used 15 pairs of NSCLC tissue samples (including cancerous tissue and para-cancerous tissues) carrying out a oligonucleotide microarray (Affymetrix GeneChip Human Transcriptome Array 2.0, Thermo Fisher Scientific) for the expression profile of NSCLC-related genes during 2014–2015 year. The sample information can be obtained from previous studies (9). Besides, there are another 43 pairs of NSCLC tissue samples for mRNA expression and protein verification during 2015–2016 year. The tissue sample information are shown in Tables S1 and S2.
The patient’s condition had been confirmed before the resection, and the clinical sample had not been made with treatment before, with the tumors in other parts of the body not metastasized to the lung. The pathological stage was determined by the Tumor Lymph Node Metastasis (TNM) staging system of the American Joint Committee on Cancer (AJCC) Clinical Practice Guidelines [2011]. The surgeon removed the excised tissue from the surgery room and fashioned it in the cryotube, then timely putting the tissue cryotube into the liquid nitrogen for cryopreservation (the tissue was within half an hour of the body). Next, the liquid nitrogen was returned to the laboratory in time. Finally, those samples were recorded basic information and stored in an ultra-low temperature freezer at −80 °C.
The MESP1 gene expression and gene ontology (GO) analysis via the Oncomine, Ualcan and DAVID online tool
The chip data of MESP1 in NSCLC was conducted with the Oncomine database (http://www.oncomine.org/) with the following filter conditions: MESP1, cancer vs. normal analysis, lung cancer, mRNA, clinical specimen. Thus, the mRNA expression data of concerning relevant NSCLC and adjacent non-tumor lung tissues (ANT) or normal tissues was collected from the Oncomine database. In addition, the expression level of MESP1 mRNA in LC can also be analyzed from the Ualcan database (http://Ualcan.path.uab.edu/). At the same time, we found that the expression of MESP1 in individual cancer stages and histological subtypes in lung adenocarcinoma (similar to lung squamous cell carcinoma) was also interesting. Then, we uploaded up-regulated genes into the filter box of DAVID with fold change (FC) ≥4.0 and P≤0.05. The functional annotation of those genes was then elucidated by GO analysis on DAVID tool.
Cell lines and siRNA transfection
The human NSCLC cell lines A549 (KCB200434YJ) and NCI-H292 (KCB2010102YJ) as well as the normal human lung epithelial cells BEAS-2B (KCB200922YJ) were purchased from Chinese Kunming Cell Bank. (Yunnan, China), and another NSCLC cell line (GLC) was obtained from 128 cancer tissues of patients from Gejiu, a region in the Yunnan Province of China. A549, NCI-H292 and BEAS-2B cells were maintained in RPMI-1640 medium (HyClone, South Logan, UT, USA). Furthermore, the GLC was cultivated in DMEM medium (HyClone, South Logan, UT, USA). All the above cells are supplemented with 10% FBS (HyClone, South Logan, UT, USA), 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA, USA) and incubated at 37 °C with 5% CO2 in incubator. Then, MESP1-targeting inhibitor siRNA (siMESP1: 5'-GCCUGGCUAUCCGCUAUAUTT-3') and a negative control siRNA (siNC: 5'-UUCUCCGAACGUGUCACGUTT-3') were synthesized by Genepharm Technologies (Shanghai, China). When the cells are in the logarithmic growth phase, and each plate of cells had reached 70–80% confluence, we transfected them into cells by Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) as carrier. Next, cells were incubated for 48 h in antibiotic-free medium for subsequent functional assays.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from NSCLC specimens or cultured cells by TRIzolTM reagent (Invitrogen, Carlsbad, CA, USA). The RNA was reverse-transcribed into cDNA by using a PrimeScript RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instructions. The concentration and purity of RNA were identified by measuring the absorbance using the NanDrop 2000C (Invitrogen, Carlsbad, CA, USA).
qRT-PCR reaction system was performed by the cDNA template with TBGreen Premix Ex TaqTM (Takara Biotechnology Co, Ltd.) (TaKaRa, Dalian, China), primer, and nuclease-free water. All reactions were performed using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with the following protocol: 95 °C for 30 s, followed by 39 cycles of 95 °C for 5 s and 60 °C for 30 s. Those data were normalized to the expression of GAPDH. The sequences of the primers (Sangon Biotech, Shanghai, China) were as follows. MESP1: forward, 5'-TGGCTCTGTTGGAGACCTGGATG-3'; reverse, 5'-CAGTCTGCCAAGGAACCACTTCG-3'. GAPDH: forward, 5'-GACAGTCAGCCGCATCTTCT-3'; reverse, 5'-TTAAAAGCAGCCCTGGTGAC-3'. The amplification specificity was confirmed by melting curve analysis. In addition, each sample was conducted in triplicate, and the mRNA expressions were calculated by the 2−ΔΔCT method.
Cell proliferation and colony formation assays
CCK-8 (Dojindo Lab, Kumamoto, Japan) and RTCA (ACEA Biosciences, San Diego, CA, USA) were used to assess the proliferation ability of cells after transfection with siMESP1 and siNC. For CCK-8 assay, each sample was incubated with 3.0×103 cells/well/100 µL in 96-well plates (Corning Costar, Corning, NY, USA) with triplicate. Then, 10 µL/well CCK-8 solution was added to each well and incubated at 37 °C for 1 h. The absorbance was calculated at a wavelength of 450 nm with the Multiskan FC microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). So the cell viability was measured at 0, 24, 48 and 72 h after transfection. For RTCA assay, it began with baseline adjustment of instrument, which added 5,000 cells/well/90 µL of into the E-Plate 16-Well (ACEA Biosciences, San Diego, CA, USA). After incubated at 37 °C for 24 h, transfection solution (well/10 µL) was added into corresponding well continuing to be cultured. Finally, cytoactive would be detected by RTCA DPlus (ACEA Biosciences, San Diego, CA, USA) until 72 h.
For colony formation assay, after transfection reaching 48 h, the cells were plated in 12-well plates (approximately 5×103 cells per well) and maintained for 3–7 days keeping in incubator at 37 °C. The medium was replaced with fresh medium periodically. We observed the appropriate colonies in the microscope, aspirating original medium, and then rinsed with PBS in a 12-well plate. Next, the clumped cell was fixed with 4% paraformaldehyde (Sangon Biotech, Shanghai, China) for 1 h. Then the colonies were stained with 0.25% crystal violet (Giemasa, Solarbio, Beijing, China) staining for 1 h. Finally, we washed plates with ddH2O, counting the number of colonies, and place them in each well taking a panorama.
Cell metastasis analysis
Cell metastasis analysis were usually consist of migration and invasion assays that were performed 24-well transwell chambers with 8 µm pores filters (Corning Costar, Corning, NY, USA) and CIM-Plate 16-well (ACEA Biosciences, San Diego, CA, USA). For transwell assay, cells were incubated with serum medium for 42 h and were starved with serum-free for 6 h in 24-well transwell chambers after transfection. Which were digested by trypsin and seeded into the upper chamber at 1.0×104 cells/well/200 µL with 100 µL of precoating matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The lower chamber was filled with 800 µL of medium with 20% FBS. All cells incubated for 48 h at 37 °C, non-metastasis cells were expelled from the upper surface with clean cottons swabs, while metastasis cells on other surface were fixed in 4% paraformaldehyde and stained with giemsa. Finally, the numbers of migrating or invading cells were counted using a Nikon TE2000 microscope (Nikon, Tokyo, Japan) in five randomly selected visual fields.
For RTCA assay, 165 µL of 20% FBS medium was added to the lower chamber in CIM-Plate 16-well after transfection. Next, we fastened the upper and lower chambers according to the operating instructions, and seeded 100 µL of serum-free matrigel medium into the upper chamber for baseline adjustment. Then, removing from original medium in upper chamber, the prepared cell suspension was seeded into upper chamber at 8×104–1×105 cells/well/100 µL. Putting the plate back to the instrument proceeded to detect 72 h. The cell migration assay was implemented in the same conditions as the invasion assay, except that the upper chamber was not coated with matrigel.
Cell cycle and apoptosis analysis
Cells were harvested after transfecting 48 h and washed by phosphate buffer saline (PBS; HyClone, South Logan, UT, USA). For cell cycle analysis, cells needed to be fixed with ice-cold 70% ethanol at 4 °C over 12 h. The fixed cells were washed with advisable PBS twice, stained with propidium iodide (PI, KeyGEN BioTECH, Nanjing, China) mixture (PI:RNase =9:1) at room temperature for avoiding light 40–60 min. DNA content was analyzed on a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with excitation wavelength of 488 nm. The percentages of the cells in three (G0/G1, S and G2/M) phases were assessed with FlowJo 7.6.1) (TreeStar Software, San Carlos, CA, USA).
For apoptosis assay, the transfected cells were digested and then double stained with Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and PI by using Annexin V-FITC apoptosis detection kit (Vazyme, Nanjing, China). Cells were mixed with 100 µL of binding buffer. Next, 5 µL Annexin V-FITC and 5 µL PI stained the samples avoiding light at room temperature 10 min. The cells were analyzed by BD FACSJazz flow cytometry within 2 h, and the data was analyzed using FlowJo 7.6.1.
Western blot analysis
Total protein of cells was obtained using lysis buffer (Biomiga, Inc., San Diego, CA, USA) and patient sample using Ripa buffer (Biomiga, Inc., San Diego, CA, USA) supplemented with protease inhibitor cocktail (Beyotime, shanghai, China) and phosphatase inhibitor cocktail (Beyotime, shanghai, China). The concentration of protein was determined by the BCA protein assay kit (Dingguochangsheng, Beijing, China). Which were denatured at high temperature by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (Takara, Dalian, China). The protein samples (15 µg) was separated on a 10% SDS-PAGE and blotted onto 0.2 µm polyvinylidene difluoride (PVDF, Millipore, Bedford, MA, USA) membranes. Then, the membranes were blocked with blocking buffers at room temperature for 1 h and incubated with appropriate primary antibodies overnight at 4 °C. After being washed by TBST 10 min three times, the membrane was incubated with secondary anti-bodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:5,000) subsequently for 1 h at room temperature. Membranes should be washed another three times for 10 min each with TBST. Finally, the proteins were visualized using a chemiluminescence reagent according to the manufacturer’s instructions (Protein Simple Jess, Bio-techne, San Diego, CA, USA). The primary antibodies include: Anti-MESP1 (ab86419, Abcam, Cambridge, USA), Anti-β-Tubulin (MA5-16308, Invitrogen, Carlsbad, USA), Anti-GAPDH(ab9485, Abcam), Anti-N-Cadherin (ab18203, Abcam), Anti-PARP1 (ab32138, Abcam), Anti-Cleaved PARP1 (ab32064, Abcam), Anti-Caspase3 (ab13585, Abcam), Anti-Active Caspase3 (ab32042, Abcam), Anti-Erk1/2 (ab17942, Abcam), P-Erk (ab65142, Abcam), Anti-Vimentin (ab137321, Abcam), Anti-Cyclin D1 (ab134175, Abcam), Anti-Cyclin B1 (ab72, Abcam), Anti-P-P38 (ab4822, Abcam). The data were normalized to GAPDH or β-Tubulin loading controls.
Statistical analysis
All statistical analyses were performed using SPSS 21.0 software (SPSS Inc., Chicago, USA). All the graphs were plotted by GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) and FlowJo7.6.1 software. All experiments were performed in triplicate by technical replicates. A t-test was used to determine differences between the experimental group and the control group. P<0.05 indicated a statistically significant difference. P*, P**, and P*** indicated significance at the 0.05, 0.01 and 0.001 levels, respectively.
Results
The expression of MESP1 was upregulated, and biological process of genes correlated with apoptosis and positive regulation of transcription in NSCLC based on bioinformatics analysis
We performed human gene expression microarray studies on 15 pairs of NSCLC and ANT. The differential expression of genes was determined by the fold change (FC) and the P-value calculated by the t-test, as was shown in the heat map (Figure 1A). We selected the MESP1 gene with P<0.05, FC =6.72. Through the Ualcan website, the profiling of available cancer genome data showed that MESP1 was highly expressed across all stages of lung adenocarcinoma (Figure 1B). And the expression of MESP1 also had a significant increase compared with the normal in histological subtypes (Figure 1C), indicating the significance of this gene in disease progression and classification. Otherwise, form the Oncomine database, we further concluded that MESP1 mRNA expression was exceedingly high in lung adenocarcinoma tissues (n=226) compared to normal tissues (n=20). What’s more, in 58 paired samples, the mRNA expression level of MESP1 was significantly higher than that in ANT (Figure 1D). In the same way, we found the same result through the Ualcan website (Figure 1E).
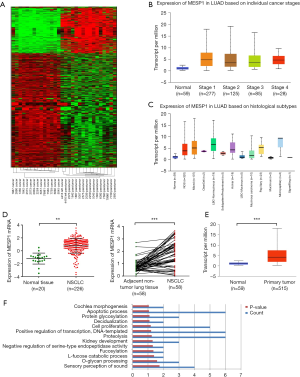
Since thresholds were set for up-regulation in cases with FC ≥4.0 and the P≤0.05. In total, there were 109 up-regulated mRNAs identified in the NSCLC samples by t-test. Based on 109 upregulated differentially mRNAs expression, an enriched map of biological processes was obtained by the functional annotations of those genes (Figure 1F, data information see Table S3). Those results showed that high expression genes were gathered during the apoptotic process. And we had found that the MESP1 stood an important position in transcription regulation. So we can conclude that it also played a key role in cell proliferation and metastasis processes.
The expression of MESP1 was upregulated in NSCLC cells and was downregulated by siMESP1 transfection
First, we examined mRNA levels of MESP1 in NSCLC, such as A549, NCI-H292, GLC and BEAS-2B cells or 4 pairs of lung squamous cell carcinoma and 39 pairs of lung adenocarcinoma (including adjacent non-tumor lung tissue and NSCLC tissue) samples by qRT-PCR. Results showed that MESP1 mRNA expression level was significantly increased in NCI-H292 and GLC cells compared to BEAS-2B. At the same time, mRNA expression of MESP1 was markedly higher in patients with NSCLC compared with adjacent nor-tumor lung tissue (Figure 2A). Next, we verified the abnormal expression of MESP1 by WB analysis at the cellular and clinical tissues, which were consistent with the above results (Figure 2B,C). And the staging levels and histological subtype classification in the patient samples were consistent with the expression changes observed in the TCGA data.
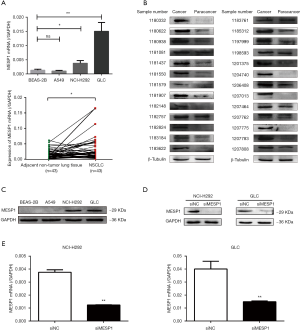
Taken together, all the results showed that MESP1 was abundant in NCI-H292 and GLC cells. The expression of MESP1 was successfully knocked down in NCI-H292 and GLC cells after transfecting siMESP1 (Figure 2D,E).
Knocking down MESP1 suppressed proliferation by blocking cell cycle progression in NSCLC cells
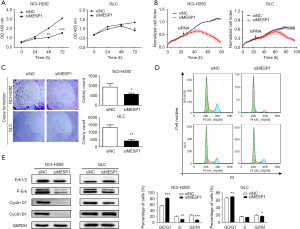
Disrupting cell cycle regulatory mechanisms can lead to cell growth uncontrolled and tumorigenesis. So, we transfected siRNA into NCI-H292 and GLC cells to suppressing the expression of endogenous MESP1. It’s observed that NCI-H292 and GLC cells displayed an effect of inhibiting cell growth activity compared to siNC group in 72 h by CCK-8 analysis (Figure 3A). Similarly, through the RTCA experiment, it’s found that the growth curve of NCI-H292 and GLC cells began to separate about 40 h. The cell index of siNC group increased continuously, while the cell activity of the siMESP1 group decreased significantly. Especially after 80 h, the treatment group has a markedly inhibitory effect (Figure 3B). In addition, the colony formation assay also indicated that the proliferation ability of NCI-H292 and GLC cells was strongly suppressed by silencing MESP1 (Figure 3C). These result showed that the down-regulation of MESP1 could dramatically repress the proliferation of NSCLC cells. Meanwhile, for flow cytometry assay, it’s depicted that the ratios of G0/G1 phase increased, but G2/M phase decreased in NCI-H292 and GLC cells (Figure 3D). Besides, the WB analysis showed that the expression of cyclin D1, cyclin B1 and p-Erk were substantially decreased (Figure 3E). So, we suspected that inhibiting the expression of MESP1 could prevent NSCLC cells from entering mitosis and DNA synthetic via Cyclin D1, Cyclin B1 and P-Erk signaling pathways.
Effect of downregulating MESP1 on the abilities of cells migration and invasion via EMT pathway
In cancer metastasis, EMT pathway was closely related to the occurrence and development of tumors. Combined with transwell assays, it’s showed that after transfection of siMESP1, the migration and invasion abilities of NCI-H292 and GLC cells were sharply weakened (Figure 4A). With the increase of time, the cell index of migration and invasion was suppressed in the treatment group (Figure 4B).
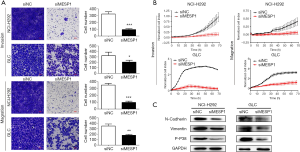
Also in the WB experiment, the expression of interstitial marker proteins such as vimentin and N-cadherin were decreased (Figure 4C). It all suggested that MESP1 promotes the progression of NSCLC by activating the EMT pathway. In addition, a variety of inflammatory factors or stress reactions can make phosphorylation in p38 MAPK, thus activating p38 MAPK (Figure 4C). P38 MAPK signal pathway was also involved in regulating apoptosis (10), proliferation, survival and metastasis (11) of cells.
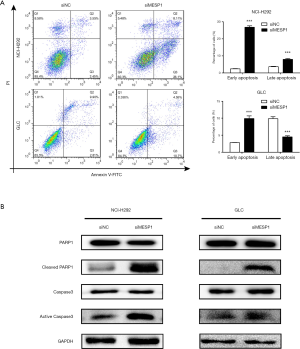
Downregulation MESP1 activated Caspase3 /PARP1 signal pathway and induced cell apoptosis
Combined with the previously biological processes analysis, the results showed that the cell cycle and apoptotic pathways were closely related to the highly expressed differential genes. So, we hoped to deeply explore the potential mechanism of apoptosis. We found that MESP1 inactivation had been shown to promote apoptosis. Annexin V-FITC/PI staining results suggested that knock-down MESP1 significantly induced apoptosis in NCI-H292 and GLC cells. The early apoptosis rate of cells in treatment group was higher than that in control group (P<0.001) (Figure 5A). Through WB experiment, we found that active Caspase3 was activated with up-regulated, and its substrate PARP1 was sheared. The expression of Cleaved PARP1 was significantly up-regulation (Figure 5B). An important substrate for Caspase3 cleavage was PARP1, a rich DNA-binding enzyme that detected signals for DNA strand breaks. Further cleavage of PARP1 showed a remarkable up-regulation of Cleaved PARP1 expression. These results suggested that the inhibition of MESP1 expression could promote apoptosis in NSCLC cells via Caspase3 /PARP1 signal pathway.
In conclusion, the inhibition of the MESP1 gene plays important regulatory roles in the development and progression of NSCLC regardless of cell proliferation, metastasis, cycle or apoptosis.
Discussion
In this paper, the high expression of the MESP1 gene was from differentially expressed genes of clinical NSCLC samples. LC-related mortality had increased more than 400% over the past three decades in China (9,12), but the treatment effects were difficult to predict. Therefore, it was important to look for predictive biomarkers to provide new insights, diagnosis and treatments. In the current study, we intended to elucidate the high expression of MESP1 in NSCLC and its effect on the growth, metastasis, cycle, and apoptosis of NSCLC cells. There was discussion about its potential molecular mechanism of action. MESP1 belongs to the family of bHLH transcription factors. As a transcription factor, MESP1 can regulate DNA replication and transcription. It was a key factor in the development of embryos in cells and is involved in cell differentiation processes including the heart (13,14), embryo (15,16). It can develop into organs such as heart, lung, liver and kidney. MESP1 played a crucial role in the age development of mesoderm-derived cardiovascular lines (17). The transcriptomic analyses of MESP1-regulated in differentiating embryoid bodies explained that gene targets of the mesoderm and endoderm lineages related to lung development were up-regulated, and it was the mediator of the earliest mesoderm progenitor cells and endoderm organogenesis (18). Many recent studies had shown that MESP1 played a key role in tumorigenesis involved in tumorigenesis, development, and metastasis (19). What’s more, there were articles reported that MESP1 had a low prognosis in LC (20). While its expression and functions were unknown in pathophysiology.
Based on the available researches, we further confirmed that MEPS1 may be a candidate for treatment or a diagnostic marker in tumor development and histological subtype classification. However, there were few studies on the signaling pathway involvement and physiological functions of MESP1. Therefore, we observed the effects of MESP1 on the growth, metastasis and apoptosis of cancer cells in NCI-H292 and GLC cells. EMT was considered the crucial step in the initial phase of the tumor metastasis cascade (21). EMT can let cancer cells acquiring invasive and metastatic characteristics, and its activation was observed in many types of cancer. More importantly, the downregulation of MESP1 could block the EMT pathway and decrease the invasion and migration of NSCLC. Similarly, in the apoptotic signaling pathway, the PARP1 was also a signal molecule for detecting DNA strand breaks (22). It’s found that down-regulating MESP1 can increase NSCLC cells degree of apoptosis via Caspase3/PARP1 signal pathway.
Conclusions
In summary, this study demonstrated that MESP1 as an oncogene was up-regulated in NSCLC cell lines and clinical tissues. In addition, siRNA-mediated silencing MESP1 had a negative regulatory effect on cell proliferation and metastasis, blocking the cell cycle and promoting cell apoptosis suggesting MESP1 as a new candidate therapeutic target for NSCLC treatment. It may provide new insight into the pathogenesis of NSCLC and improve its clinical prognosis. Therefore, the specific role of MESP1 in the occurrence and progression of NSCLC needed to be further studied.
Table S1
| Sample number | Gender | Age | Tumor stage (TNM) | Clinical stages |
|---|---|---|---|---|
| 1 | Female | 39 | T1N0M0 | IA |
| 2 | Female | 43 | T2aN1M0 | IIA |
| 3 | Female | 43 | T3N0M0 | IIB |
| 4 | Male | 44 | T1N0M0 | IA |
| 5 | Female | 50 | T1N0M0 | IA |
| 6 | Female | 51 | T1N0M0 | IA |
| 7 | Male | 51 | T2aN1M0 | IIA |
| 8 | Male | 52 | T2aN0M0 | IB |
| 9 | Male | 54 | T1N0M0 | IA |
| 10 | Male | 58 | T2aN0M0 | IB |
| 11 | Male | 60 | T2bN2M0 | IIIA |
| 12 | Male | 62 | T1N0M0 | IA |
| 13 | Female | 63 | T1bN0M0 | IA |
| 14 | Male | 65 | T1aN0M0 | IA |
| 15 | Female | 69 | T1N0M0 | IA |
NSCLC, non-small cell lung cancer.
Table S2
| Sample number | Gender | Age | Tumor stage (TNM) | Clinical stages |
|---|---|---|---|---|
| 1 | Male | 42 | T1aN0M0 | IA |
| 2 | Male | 42 | T2aN0M0 | IB |
| 3 | Male | 51 | T4N0M0 | IIIA |
| 4 | Female | 52 | T1N0M0 | IA |
| 5 | Male | 38 | T2aN1M0 | IIB |
| 6 | Male | 45 | T1bN0M0 | IA |
| 7 | Female | 58 | T1aN0M0 | IA |
| 8 | Male | 53 | T3N0M0 | IIB |
| 9 | Female | 54 | T1N0M0 | IA |
| 10 | Male | 75 | T2N0M0 | IB |
| 11 | Male | 71 | T1N0M0 | IA |
| 12 | Male | 65 | T1bN2M1a | IV |
| 13 | Male | 54 | T1bN2M1 | IV |
| 14 | Male | 66 | T2N2M1 | IV |
| 15 | Female | 41 | T1bN0M0 | IA |
| 16 | Male | 39 | T2aN0M0 | IB |
| 17 | Female | 60 | T1bN2M0 | IIIA |
| 18 | Male | 46 | T1bN0M0 | IA |
| 19 | Male | 62 | T1aN0M0 | IA |
| 20 | Male | 61 | T1bN0M0 | IA |
| 21 | Male | 65 | T1aN0M0 | IA |
| 22 | Female | 68 | T2aN2M0 | IIIA |
| 23 | Male | 68 | T1bN1M0 | IIB |
| 24 | Male | 53 | T2N2M1 | IV |
| 25 | Male | 69 | T1bN0M0 | IA |
| 26 | Female | 53 | T2N2M0 | IIIA |
| 27 | Male | 43 | T1aN0M0 | IA |
| 28 | Male | 51 | T1bN2M0 | IIIA |
| 29 | Male | 66 | T1aN0M0 | IA |
| 30 | Female | 44 | T1BN2M | IV |
| 31 | Female | 56 | T1bN2M0 | IIIA |
| 32 | Male | 75 | T1bN0M0 | IA |
| 33 | Female | 50 | T1aN0M0 | IA |
| 34 | Male | 67 | T4N2M0 | IIIB |
| 35 | Male | 41 | T1bN2M0 | IIIA |
| 36 | Male | 62 | T1bN0M0 | IA |
| 37 | Female | 51 | T1N0M0 | IA |
| 38 | Male | 48 | T1aN0M0 | IA |
| 39 | Male | 59 | T4N2M1 | IV |
| 40 | Male | 54 | T1aN0M0 | IA |
| 41 | Female | 53 | T1bN0M0 | IA |
| 42 | Female | 53 | T1aN0M0 | IA |
| 43 | Male | 66 | T2aN0M0 | IA |
Note: TNM staging of tumor is in accordance with NCCN revised NSCLC staging standard in 2011. NSCLC, non-small cell lung cancer.
Table S3
| Category | Term | Count | P value | Genes | List total | Pop hits | Pop total | Fold enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | Sensory perception of sound | 4 | 1.742712 | SIX1, TFAP2A, COL1A1, GJB2 | 71 | 133 | 16,792 | 7.112994 | 0.999909 | 0.999909 | 23.1664 |
| GOTERM_BP_DIRECT | O-glycan processing | 3 | 1.585782 | GCNT3, MUC21, B3GNT6 | 71 | 60 | 16,792 | 11.82535 | 0.999999 | 0.998776 | 31.59682 |
| GOTERM_BP_DIRECT | L-fucose catabolic process | 2 | 1.388023 | FUT3, FUT2 | 71 | 10 | 16,792 | 47.30141 | 1 | 0.999178 | 45.30483 |
| GOTERM_BP_DIRECT | Fucosylation | 2 | 1.347518 | FUT3, FUT2 | 71 | 11 | 16,792 | 43.00128 | 1 | 0.99715 | 48.50853 |
| GOTERM_BP_DIRECT | Negative regulation of serine-type endopeptidase activity | 2 | 1.310617 | SPINK1, SPINK5 | 71 | 12 | 16,792 | 39.41784 | 1 | 0.993993 | 51.52475 |
| GOTERM_BP_DIRECT | Kidney development | 3 | 1.300744 | PKHD1, SIX1, TFAP2A | 71 | 86 | 16,792 | 8.250246 | 1 | 0.987258 | 52.34588 |
| GOTERM_BP_DIRECT | Proteolysis | 6 | 1.241403 | NRIP3, UNC5CL, WFDC2, MMP11, TMPRSS4, HABP2 | 71 | 500 | 16,792 | 2.838085 | 1 | 0.98648 | 57.386 |
| GOTERM_BP_DIRECT | Positive regulation of transcription, DNA-templated | 6 | 1.197338 | CDKN2A, SIX1, TFAP2A, COL1A1, MESP1, TOX3 | 71 | 515 | 16,792 | 2.755422 | 1 | 0.984721 | 61.21472 |
| GOTERM_BP_DIRECT | Cell proliferation | 5 | 1.177723 | STIL, FAM83A, DLGAP5, SIX1, MELK | 71 | 366 | 16,792 | 3.230971 | 1 | 0.979645 | 62.93233 |
| GOTERM_BP_DIRECT | Decidualization | 2 | 1.095847 | SPP1, GJB2 | 71 | 20 | 16,792 | 23.6507 | 1 | 0.985925 | 70.09516 |
| GOTERM_BP_DIRECT | Protein glycosylation | 3 | 1.093127 | B3GNT6, FUT3, FUT2 | 71 | 113 | 16,792 | 6.278948 | 1 | 0.979782 | 70.33085 |
| GOTERM_BP_DIRECT | Apoptotic process | 6 | 1.058088 | CDKN2A, CDCA7, SIX1, TOX3, PHLDA2, MELK | 71 | 567 | 16,792 | 2.50272 | 1 | 0.979569 | 73.33903 |
| GOTERM_BP_DIRECT | Cochlea morphogenesis | 2 | 1.056218 | CTHRC1, SIX1 | 71 | 22 | 16,792 | 21.50064 | 1 | 0.972884 | 73.49784 |
Acknowledgments
Funding: This project was supported by
Footnote
Reporting Checklist: The authors have completed the CONSORT reporting checklist and MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1132
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1132
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure (available at http://dx.doi.org/10.21037/tcr-20-1132). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was signed by every patient and this experiment gained permission from ethics committee of the Yunnan Cancer Hospital (approval number: KY202014).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev 2015;41:361-75. [Crossref] [PubMed]
- Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Eskildsen TV, Ayoubi S, Thomassen M, et al. MESP1 knock-down in human iPSC attenuates early vascular progenitor cell differentiation after completed primitive streak specification. Dev Biol 2019;445:1-7. [Crossref] [PubMed]
- Lindsley RC, Gill JG, Murphy TL, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 2008;3:55-68. [Crossref] [PubMed]
- Werner P, Latney B, Deardorff MA, et al. MESP1 Mutations in Patients with Congenital Heart Defects. Hum Mutat 2016;37:308-14. [Crossref] [PubMed]
- Li Q, Ran P, Zhang X, et al. Downregulation of N-Acetylglucosaminyltransferase GCNT3 by miR-302b-3p Decreases Non-Small Cell Lung Cancer (NSCLC) Cell Proliferation, Migration and Invasion. Cell Physiol Biochem 2018;50:987-1004. [Crossref] [PubMed]
- Gao X, Li N, Zhang J. SB203580, a p38MAPK inhibitor, attenuates olfactory dysfunction by inhibiting OSN apoptosis in AR mice (activation and involvement of the p38 mitogen-activated protein kinase in olfactory sensory neuronal apoptosis of OVA-induced allergic rhinitis). Brain Behav 2019;9:e01295. [Crossref] [PubMed]
- Zhang L, Huang D, Shao D, et al. Fenretinide inhibits the proliferation and migration of human liver cancer HepG2 cells by downregulating the activation of myosin light chain kinase through the p38MAPK signaling pathway. Oncol Rep 2018;40:518-26. [PubMed]
- Fu M, Fan W, Pu X, et al. Elevated expression of SHIP2 correlates with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 2013;6:2185-91. [PubMed]
- den Hartogh SC, Wolstencroft K, Mummery CL, et al. A comprehensive gene expression analysis at sequential stages of in vitro cardiac differentiation from isolated MESP1-expressing-mesoderm progenitors. Sci Rep 2016;6:19386. [Crossref] [PubMed]
- Lescroart F, Wang X, Lin X, et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 2018;359:1177-81. [Crossref] [PubMed]
- Wang H, Zhao S, Barton M, et al. Reciprocity of Action of Increasing Oct4 and Repressing p53 in Transdifferentiation of Mouse Embryonic Fibroblasts into Cardiac Myocytes. Cell Reprogram 2018;20:27-37. [Crossref] [PubMed]
- Sun J, Ting MC, Ishii M, et al. Msx1 and Msx2 function together in the regulation of primordial germ cell migration in the mouse. Dev Biol 2016;417:11-24. [Crossref] [PubMed]
- Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res 2010;107:1414-27. [Crossref] [PubMed]
- Soibam B, Benham A, Kim J, et al. Genome-Wide Identification of MESP1 Targets Demonstrates Primary Regulation Over Mesendoderm Gene Activity. Stem Cells 2015;33:3254-65. [Crossref] [PubMed]
- Chiapparo G, Lin X, Lescroart F, et al. Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J Cell Biol 2016;213:463-77. [Crossref] [PubMed]
- Tandon N, Goller K, Wang F, et al. Aberrant expression of embryonic mesendoderm factor MESP1 promotes tumorigenesis. EBioMedicine 2019;50:55-66. [Crossref] [PubMed]
- Dominguez C, David JM, Palena C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin. Cancer Biol 2017;47:177-84. [Crossref] [PubMed]
- Decker P, Muller S. Modulating poly (ADP-ribose) polymerase activity: potential for the prevention and therapy of pathogenic situations involving DNA damage and oxidative stress. Curr Pharm Biotechnol 2002;3:275-83. [Crossref] [PubMed]


