
FAM172A affects cell proliferation and apoptosis not by targeting β-tubulin in HepG2 cells
Introduction
The current two first-line systemic treatment drugs for liver cancer, sorafenib, and lovastinib can extend the patient’s median survival time by about 1 year and accordingly extend the patient’s overall survival time (1,2). However, this prognosis is far from meeting clinical expectations, and there is an urgent need to find more effective treatments and biological targets. Owing to the outstanding effect of tubulin in cell division and cell cycle regulation (3-7), tubulin can be a potential target for the development of new anti-tumor drugs (8,9). Studies have found that tubulin affects the proliferation and migration of liver cancer cell lines between Huh7 and HepG2 (10,11).
The system with sequence similarity 172 and member A (FAM172A or C5ORF21) is a human protein encoded by the FAM172A gene. After the complete human genome was sequenced in 2001, the gene was initially found from the collection of human cDNA libraries (12). Li et al. cloned the FAM172A gene from normal human aortic tissues. They showed that high glucose levels up-regulated the expression of FAM172A protein in human aortic smooth muscle cells in a concentration- and time-dependent manner and speculated that FAM172A as a new Proteins might be involved in the pathogenesis of high glucose-induced vascular injury (13,14). Chunhui Cui et al. found that FAM172A can inhibit the proliferation and invasion ability of colorectal cancer cell line LOVO cells and infer that this gene may be a tumor suppressor for colorectal cancer (15). The results of Qian et al. showed that the FAM172A gene could inhibit the proliferation of colorectal cancer cells and promote apoptosis and differentiation, and the transcription factor STAT1 can bind to the promoter region of the FAM172A gene and up-regulate the expression of this gene (16).
To date, whether FAM172A affects the biological behavior of liver cancer cells by acting on tubulin has not been reported. This work will explore the potential interaction between FAM172A and tubulin, and the biological impact on hepatocellular carcinoma cells. This research can supply targets and a theoretical basis for liver cancer treatment and the development of new drugs. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2868).
Methods
Study cell line
The human Liver cancer HepG2 cell line was from the Second People’s Hospital of Guangdong Province. The cells were verified by short tandem repeat (STR) testing. The cells were cultured with the Dulbecco’s modified eagle medium (11885092; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS, CCS30013.01HI, MRC, Changzhou, China), supplemented with 1% penicillin-streptomycin (10,000 U/mL) (15140122; Gibco) and incubated at 37 °C with 5% CO2.
The study was approved by the Education and Research Committee and the Ethics Committee of The First People’s Hospital of Kashgar. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013), and written informed consent was obtained from the participants.
Lentiviral vector FAM172A Transfection
Shanghai GeneChem Co., Ltd constructed the FAM172A lentiviral expression vectors. The lentiviral expression vectors of PLVX-mCMV-ZsGreen-PGK-Puro-homo-FAM172A were transfected into the HepG2 cells using Polybrene (Shanghai GeneChem Co., Ltd., China) and Enhanced Infection Solution (Shanghai GeneChem Co., Ltd., China), at the multiplicities of infection of 50, 10 and 20, respectively. The blank and scrambled small interfering RNA (siRNA) negative control groups were also set up. Shanghai GeneChem Co., Ltd. synthesized the siRNA; however, the sequences were not available. Following the addition of the lentiviral vectors, the cells were incubated at 37 °C in a 5% CO2 incubator for 16 h. Subsequently, the cells were cultured with fresh DMEM for 72 h before collection.
Cell viability
The cells were seeded onto the 96-well plates, at a density of 1×105 cells/mL, in 180 µL culture medium each well. After cultured in a 37 °C, 5%CO2 incubator for 24 h, the cells were subjected to different treatments, respectively, for 7 days. Then 20 µL MTT solution (M1020; Solarbio, Wuhan, Hubei, China) was added into each well to incubate the cells at 37 °C for 4 h. The absorbance at 450 nm was added with the Multiskan GO microplate reader (Thermo, USA). Each group had 5 replicate wells, and the cell viabilities were calculated.
Cellular immunofluorescence
The HepG2 were seeded onto the 96-well plates, at a density of 1×105 cells/ml, in 100 µL culture medium each well. After intervention under different conditions, the medium was aspirated, and the cells were gently washed with PBS for 3 times. Totally 30 µL paraformaldehyde (4%) was added to each well to fix the cells for 15 min, followed by another round of washing with PBS for 3 times. After treated with 0.5% Triton X-100 at room temperature for 20 min and the following washing, the cells were blocked with 5% BSA solution at room temperature for 20 min. Then the cells were incubated with the rabbit anti -β-tublin primary antibody (1:1,000 dilution; bs-10680R; Bioss), at 4 °C overnight. After washing, the cells were incubated with fluorescent secondary antibody (1:500 dilution; bs-0295G-FITC; Bioss) at 37 °C in the dark for 2 h. After washing, the cells were incubated with DAPI (C0060; Solarbio) in the dark for 5 min. Fluorescence was observed with an inverted fluorescence microscope (Axio Observer A1, Zeiss, Germany), and the images were taken and analyzed.
Western blot analysis
The HepG2 cells were lysed using Biosharp lysis (BL504A). The protein concentration was determined using the bicinchoninic acid assay method. Protein samples (40 µg per lane) were loaded onto a 4% gel, resolved SDS-PAGE, and then later transferred onto a PVDF membrane. After blocking with 5% BSA at room temperature for 1 h, the membrane was incubated with anti-FAM172A (1:100; ab121364; abcam) and anti-GAPDH (1:5,000; bs-50549R; BIOSS) primary antibodies at 4 °C for 16 h. Subsequently, the membranes were incubated with the secondary antibody of goat anti-rabbit HRP conjugated IgG (1:10,000; ZB-2301; OriGene Technologies, Inc., MD, USA) at 37 °C for 1 h in the dark. The blot was developed using the ECL method (PE0010; Beijing Solarbio Science & Technology Co., Ltd., China). Protein bands were imaged and analyzed using ImageJ software version 1.52s (National Institutes of Health).
Reverse transcription-quantitative (RT-q) PCR.
Total RNA was extracted from HepG2 Cell samples using a Total RNA extraction kit (DP431; Tiangen Biotech Co., Ltd., China). The mRNAs of FAM172A and β-tublin were obtained from the GenBank database. The ORFs were designed for the PCR primers. β-tubulin: F: 5’-CCGAGAGGGAAAGTCATAACAAC-3’, R: 5’-CCAGGGT ATCAG GAAAATCAAGAG-3’. FAM172A: F: 5’-TGGCACCCAGCACAATGAA-3’, R: 5’-CTAAGTCATAGTCCGCCTAGAAGCA-3’. The extracted RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (cat. no. RR037A; Takara Bio, Inc., Shiga, Japan). The reaction conditions were 37 °C for 15 min and 85 °C for 5 sec. RT-qPCR was performed using an AGS PCR machine (AFD4800; Hangzhou AnYu Technologies Co., Ltd., China). The reaction conditions were: 95 °C for 2 min, 95 °C for 5 sec, 60 °C for 10 sec, and 72 for 30. In total, there were 40 cycles. Target gene expression levels were determined with the 2−ΔΔCqmethod. β-actin was used as an endogenous control.
Statistical analysis
The experiments were performed three times independently. Data are presented as the mean ± SD. SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. One-way ANOVA was performed for the comparison of mean values among groups followed by Tukey’s post hoc test. A χ2 test was used to compare the hypermutation ratio. P<0.05 was considered to indicate a statistically significant difference.
Results
Study on the interaction between FAM172A and β-tubulin in human liver cancer cell lines
Verification of the interaction between FAM172A and β-tubulin
In this work, the STRING 10 database was used to predict the interaction proteins of FAM172A protein. And 10 proteins, including PLAC8L1, GRIK3, and PCYOX1, which are the interaction proteins with FAM172A, were predicted (Figure 1A). In the predicted proteins by STRING, there is no β-tubulin protein, which indicates that FAM172A protein may not interact with the β-tubulin protein.
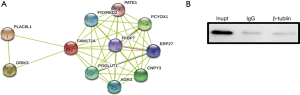
The co-immunoprecipitation method was repeated 3 times with the corresponding IgG as a control, and all showed no interaction between the two proteins (Figure 1B).
Effect of FAM172A on β-tubulin
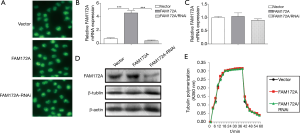
Transfect HepG2 cells with PcDNATM3.1/myc-His(−) empty vector plasmid, PcDNATM3.1/myc-His(−)-FAM172A, RNAi not related to β-tubulin and pSuper-retro-puro/fa172a/RNAi. After 48 hours, the plasmid vector was fixed, and the cells were subjected to cellular immunofluorescence detection. As shown in Figure 2A,B, there is no significant difference in the distribution of tubulin between control, FAM172A, unrelated interference, and FAM172A-RNAi groups (P>0.05). Real-time quantitative PCR detected the effect of FAM172A expression on the relative expression of the β-tubulin gene, and there is no significant difference among the treatment groups (P>0.05) (Figure 2C). Western blot results (Figure 2D) show that the FAM172A protein expression level in the FAM172A overexpression group increases significantly (P<0.05), while the FAM172A protein expression level in the interference group decreased significantly (P<0.05). The expression level of β-tubulin protein in each treatment group did not reach a significant level (P>0.05). In the tubulin polymerization-depolymerization experiment (Figure 2E), the polymerization of tubulin showed an upward trend with the increase of the reaction time at 37 °C polymerization, and the polymerization was strong within the first 20 minutes. Afterward, depolymerization occurs under 4 °C ice bath conditions, and a large amount of tubulin depolymerizes just after an ice bath. In tubulin polymerization-depolymerization, there was no significant difference among the treatment groups (P>0.05).
The effect of FAM172A on the biological function of liver cancer cells
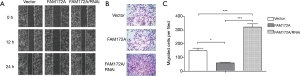
To further explore the role of FAM172A in the migration of hepatoma carcinoma cells, we used HepG2 cells to establish FAM172A overexpression and interference cells. The cell migration and invasion test results showed (Figure 3) that the upregulation of FAM172A inhibited the cell migration and invasion ability (P<0.05), and the downregulation of FAM172A, cell migration, and invasion ability was significantly enhanced (P<0.05).
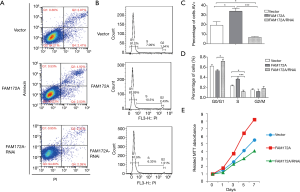
Flow cytometry and MTT methods were used to detect the effect of FAM172A on apoptosis, cell cycle, and proliferation of hepatoma carcinoma cells. The detection results of the number of apoptotic cells in each treatment group showed that the number of apoptotic cells in the FAM172A overexpression group was significantly higher than that in the other 2 groups (P<0.05). In comparison, the number of apoptotic cells in the FAM172A interference group was significantly lower than that in the other groups (P<0.05). The periodic detection results of hepatoma carcinoma cells showed that FAM172A overexpression cells increased in S period, significantly higher than the control group (P<0.05); while FAM172A interference group cells decreased in S period, smaller than the control group (P<0.05) (Figure 4A,B,C,D). With the extension of the incubation time, the numbers of hepatoma carcinoma cells in each treatment group showed an increasing trend. Also, the number of hepatoma carcinoma cells in the FAM172A interference group was more than the other 2 groups. Further, the breeding rate accelerated after 3 days of incubation (P<0.05). The number of cell breeding was the lowest in the FAM172A overexpression group, and the number of cells in the late stage of incubation was significantly lower than that of the other groups (P<0.05) (Figure 4E).
Discussion
Hepatocellular carcinoma is one of the most common human cancers in the world, and its high mortality rate is due to there are no apparent symptoms until late (17). Various environmental risk factors, including hepatitis B virus or hepatitis C virus infection, heavy drinking, dietary aflatoxin, and exposure to carcinogenic chemicals, have been found as the causes of liver cancer (18,19). However, in the past two decades, the overall survival rate of patients with hepatocellular carcinoma has not improved significantly, and the mechanism of liver cancer development has not been explained in detail (20). At present, molecular targeted therapy for advanced hepatocellular carcinoma shows certain prospects (21).
Tubulin is the current target of anti-tumor drugs: the realization of microtubule function depends on the dynamic polymerization and depolymerization process of tubulin, which provides inhibitors with tubulin to destroy the normal function of microtubules. Favorable entry point (22,23). This study intends to investigate whether FAM172A is a novel tubulin inhibitor that affects the biological function of liver cancer cells. Through bioinformatics and co-immunoprecipitation methods, it is concluded that there is no interaction between FAM172A and β-tubulin. Subsequently, it was detected by immunofluorescence experiments that the change of FAM172A expression would not affect the morphology and distribution of Tubulin in liver cancer cells. Further, qRT-PCR, Western blot and Tubulin polymerization-depolymerization experiments have shown that FAM172A is up-regulated or down-regulated. Meanwhile, β-tubulin mRNA, protein expression, and tubulin polymerization-depolymerization process will not change significantly. The above results prove that there is no targeting relationship between FAM172A and tubulin.
Regarding the role of FAM172A in cancer, there are significant differences between the results of numerous studies. In the study of the physiological function of FAM172A protein, it was found that FAM172A can promote the growth rate and cell proliferation of human embryonic kidney cells, and at the same time reduce the number of apoptosis and increase the number of cells in S phase, thus indicating that FAM172A is involved in the regulation of cell growth (24). With the development of tumors, metastasis, and invasion of blood vessels, the number of mesenchymal circulating tumor cells increased significantly. The expression of FAM172A in these tumor cells was significantly higher than that of epithelial circulating cells, showing that FAM172A is related to the degree of tumor lesions. Circulating tumor cells and FAM172A detection can predict the high-risk stage of colorectal cancer (25). miR-27a is highly expressed in colorectal cancer patients and colorectal cancer cell lines, and its expression level is positively correlated with the difference in the overall survival rate of colorectal cancer patients. Increasing or inhibiting miR-27a expression can promote or inhibit colorectal cancer cell lines Migration; dual-luciferase experiments confirmed that miR-27a directly targets the 3’-UTR of FAM172A mRNA, increasing or inhibiting the expression of miR-27a can down-regulate or up-regulate the expression of FAM172A. Therefore, by inhibiting miR-27a expression or Treatments that increase FAM172A expression prevent colorectal cancer metastases (26). The expression of FAM172A protein in papillary thyroid carcinoma tissue is significantly higher than that in adjacent tissues and normal thyroid tissue. Overexpression of FAM172A will activate the p38 MAPK signaling pathway and accelerate the proliferation of IHH-4 cells (27). This study proved through cytology that FAM172A could not only inhibit the migration, invasion, and proliferation of liver cancer cells but also promote apoptosis, causing cell cycle arrest in the S phase.
The results obtained in this study prove that FAM172A can inhibit the proliferation of liver cancer cells, but it is not achieved by interacting with β-tubulin. The molecular mechanism of FAM172A causing apoptosis of liver cancer cells and the proliferation of inhibiting cancer cells is not yet precise. Our laboratory intends to study further the mechanism and pathways of FAM172A regulating tumor cells in the network
Acknowledgments
Thanks to Xinjiang Dingju Biotechnology Co., Ltd. for their help in the experiment.
Funding: The present study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2868
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2868
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2868). The authors have no conflicts of interest to declare.
(English language editor: J. Chapnick)
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Education and Research Committee and the Ethics Committee of The First People’s Hospital of Kashgar. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013), and written informed consent was obtained from the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163-73. [Crossref] [PubMed]
- Boland P, Wu J. Systemic therapy for hepatocellular carcinoma: beyond sorafenib. Chin Clin Oncol 2018;7:50. [Crossref] [PubMed]
- Chen YC, Lu PH, Pan SL, et al. Quinolone analogue inhibits tubulin polymerization and induces apoptosis via Cdk1-involved signaling pathways. Biochem Pharmacol 2007;74:10-9. [Crossref] [PubMed]
- Hou S, Hua L, Wang W, et al. Nucleolar spindle associated protein 1 (NUSAP1) facilitates proliferation of hepatocellular carcinoma cells. Transl Cancer Res 2019;8:2113-20. [Crossref]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol 2001;3:E28-E34. [Crossref] [PubMed]
- Rusan NM, Fagerstrom CJ, Yvon AM, et al. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol Biol Cell 2001;12:971-80. [Crossref] [PubMed]
- Zhai Y, Kronebusch PJ, Simon PM, et al. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol 1996;135:201-14. [Crossref] [PubMed]
- Giannakakou P, Sackett D, Fojo T. Tubulin/microtubules: still a promising target for new chemotherapeutic agents. J Natl Cancer Inst 2000;92:182-3. [Crossref] [PubMed]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253-65. [Crossref] [PubMed]
- Rein-Fischboeck L, Pohl R, Haberl EM, et al. Tubulin alpha 8 is expressed in hepatic stellate cells and is induced in transformed hepatocytes. Mol Cell Biochem 2017;428:161-70. [Crossref] [PubMed]
- Yan W, Xiangyu C, Ya L, et al. An orally antitumor chalcone hybrid inhibited HepG2 cells growth and migration as the tubulin binding agent. Invest New Drugs 2019;37:784-90. [Crossref] [PubMed]
- Wiemann S, Weil B, Wellenreuther R, et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res 2001;11:422-35. [Crossref] [PubMed]
- Li LX, Tao Z, Dong XH, et al. Molecular cloning of a novel gene, C5orf21 gene and its roles in diabetic macroangiopathy. Zhonghua Yi Xue Za Zhi 2009;89:2574-7. [PubMed]
- Li L, Dong X, Leong MC, et al. Identification of the novel protein FAM172A, and its up-regulation by high glucose in human aortic smooth muscle cells. Int J Mol Med 2010;26:483-90. [PubMed]
- Cui C, Ye L, Huang Z, et al. FAM172A is a tumor suppressor in colorectal carcinoma. Tumour Biol 2016;37:6501-10. [Crossref] [PubMed]
- Qian K, Zhang J, Lu J, et al. FAM172A modulates apoptosis and proliferation of colon cancer cells via STAT1 binding to its promoter. Oncol Rep 2016;35:1273-80. [Crossref] [PubMed]
- Cheung TT. Management of hepatocellular carcinoma: from bench to bedside and beyond. Transl Gastroenterol Hepatol 2019;4:54. [Crossref] [PubMed]
- Serio I, Napoli L, Leoni S, et al. Direct antiviral agents for HCV infection and hepatocellular carcinoma: facts and FADs. Transl Cancer Res 2019;8:S223-32. [Crossref]
- Guarino M, Dufour JF. Smoking favours hepatocellular carcinoma. Ann Transl Med 2019;7:S99. [Crossref] [PubMed]
- Bruix J, Boix L, Sala M, et al. Focus on hepatocellular carcinoma. Cancer Cell 2004;5:215-9. [Crossref] [PubMed]
- Shen YC, Hsu C, Cheng AL. Molecular targeted therapy for advanced hepatocellular carcinoma: current status and future perspectives. J Gastroenterol 2010;45:794-807. [Crossref] [PubMed]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253-65. [Crossref] [PubMed]
- Tischfield MA, Cederquist GY, Gupta ML, et al. Phenotypic spectrum of the Tubulin-related Disorders and Functional Implications of Disease-causing Mutations. Curr Opin Genet Dev 2011;21:286-94. [Crossref] [PubMed]
- Li LX, Zhou WB, Tao Z, et al. Effect of FAM172A protein on apoptosis and proliferation in HEK293 cells. Zhonghua Yi Xue Za Zhi 2010;90:2424-7. [PubMed]
- Cui CH, Chen RH, Zhai DY, et al. Detection of FAM172A expressed in circulating tumor cells is a feasible method to predict high-risk subgroups of colorectal cancer. Tumour Biol 2017;39:1010428317699126. [Crossref] [PubMed]
- Liu W, Qian K, Wei X, et al. miR 27a promotes proliferation, migration, and invasion of colorectal cancer by targeting FAM172A and acts as a diagnostic and prognostic biomarker. Oncol Rep 2017;37:3554-64. [Crossref] [PubMed]
- Li MF, Zhang R, Guo MG, et al. FAM172A protein promotes the proliferation of human papillary thyroid carcinoma cells via the p38 mitogen-activated protein kinase pathway. Mol Med Rep 2016;13:353-8. [Crossref] [PubMed]


