MicroRNA-153-3p sensitizes melanoma cells to dacarbazine by suppressing ATG5-mediated autophagy and apoptosis
Introduction
Melanoma is a malignant tumor deriving from melanocytes, and has a high mortality rate (1). In the USA, melanoma was considered to account for 5% of all cancers in 2013, which indicates its high prevalence (2,3). Although in China the incidence of melanoma is not such high as that of the USA, the increasing morbidity and mortality rates over past years have attracted widespread concern (4). In the early stage, melanoma can be surgically removed, and the 5-year survival rate of melanoma patients is as high as 98% (5). Nevertheless, melanoma can easily metastasize to regional lymph nodes and result in poor prognosis, with a 5-year survival rate of only 16% (6,7). Various methods are used for the treatment of melanoma, including surgery, chemotherapy, radiation therapy, and immunotherapy (8,9). Dacarbazine is one of the most commonly used chemotherapeutic agents for the treatment of melanoma (10); however, only 5–10% of patients benefit from this treatment (11). The main reason is the drug resistance of dacarbazine due to various reasons including hypoxia exposure (12), BRAFV600E targeting (13), advanced melanoma (14). Melanoma is still a malignant tumor that is insensitive to traditional treatment methods such as chemotherapy drugs, so it is urgent to explore new treatments (15).
Autophagy is a self-cannibalization process that occurs in both normal and cancer cells. The cells reuse damaged proteins and organelles by autophagosomes to maintain cell homeostasis (16). Autophagy plays a dual role in cancer development and treatment. Previous studies have reported that autophagy is linked to chemotherapy resistance in cancer cells (17,18). In vivo experiments identify autophagy ATG7 to be essential for autophagy activation. Mechanistically, inhibition of the autophagic flux in dormant breast cancer cells leads to the accumulation of damaged mitochondria and reactive oxygen species (ROS), resulting in cell apoptosis (19). The inhibition of apoptosis enables the survival and proliferation of tumors and contributes to resistance to conventional chemotherapy agents (20). However, the mechanism underlying the relationship between autophagy and dacarbazine resistance in melanoma cells remains unclear and requires further elucidation.
A group of small non-coding RNAs with 20–22 of nucleotides are referred to as microRNAs (miRNAs). miRNAscan post-transcriptionally adjust various target genes by targeting their 3'-untranslated region (3'-UTR) (21). Previous research has shown that that miR-153-3p suppresses cell proliferation and invasion by downregulating Snail family transcriptional repressor-1 (SNAI1) in melanoma (22). miR-153-3p restrains autophagy and intervertebral disc degeneration by downregulating the expression of autophagy related gene 5 (ATG5) (23). More importantly, long intergenic noncoding RNA 00641 (LINC00641) induces autophagic cell death by regulating miR-153-3p. Previous studies have demonstrated that some miRNAs are vital mediators of chemoresistance in cancer cells (24). Sun et al. reported that the downregulation of miR-374b-5p promotes chemotherapeutic resistance by upregulating multiple anti-apoptotic proteins in pancreatic cancer (25). miR-34a was reported to decrease chemoresistance in ovarian cancer cells by targeting histone deacetylase-1 (HDAC1) (26). Importantly, the miR-31-SOX10 axis regulates tumor growth and dacarbazine resistance of melanoma via PI3K/AKT pathway (27). These results demonstrate that the dysregulation of miRNAs has an important role in the development of cancer cell chemoresistance.
In the present study, we explored the mechanism of miR-153-3p in the regulation of dacarbazine resistance of melanoma cells. The result revealed that miR-153-3p overexpression sensitizes melanoma cells to dacarbazine by suppressing ATG5-mediated autophagy and apoptosis, suggesting that targeting miR-153-3p may be a novel therapeutic strategy to treat melanoma patients.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2660).
Methods
Cell culture and reagents
Normal human melanocytes (HEMn-LP) and melanoma cell lines (A357 and M14) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured with RPMI 1640 media (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. Dacarbazine was obtained from Sigma and dissolved in dimethyl sulfoxide. The dose of dacarbazine were selected according to this reference (28). Antibodies used in the present study were: anti-Beclin1 (#3738, 1:1,000), anti-LC3 I/II (#12741, 1:1,000), anti-ATG5 (#12994, 1:1,000), and anti-GAPDH (#5174, 1:1,000). All antibodies were obtained from Cell Signaling Technology.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. A RT kit (RR036A; Takara Biotechnology, Dalian, China) was used for cDNA production, cDNA amplification was detected using the SYBR Premix Ex Taq GC kit (Takara Biotechnology, Dalian, China), and GAPDH was used for normalizing. The relative level was calculated by relative quantification method.
Cell transfection
miR-153-3p mimic, miR-NC, and pcDNA3.1-ATG5 were designed and synthesized by GenePharma. Transfections were performed using the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions.
Cell viability assay
Cell viability was detected by Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Shanghai, China). Cells were added into 96-well plates and treated with different concentrations of dacarbazine for 24 hours. The culture medium was then replaced in each well with 100 µL fresh medium containing 10% CCK-8 reaction solution and incubated for 2 hours. Absorbance were measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Carlsbad, CA, USA).
Flow cytometric analysis of apoptosis
9 The cell apoptotic rate was determined using the annexin V-fluorescein isothiocyanate/propidium iodide (PI) apoptosis detection kit (Multisciences). After washing with ice-cold phosphate-buffered saline three times, the cells were resuspended and incubated with 5 µL annexin V-FITC and 10 µL PI. A flow cytometer (BD Biosciences, US) was used to analyze the cell apoptotic rate. The flow cytometry excitation wavelength was 488 nm; a bandpass filter with a wavelength of 515 nm was used to detect FITC fluorescence, and another filter with a wavelength >560 nm was used to detect PI.
Immunofluorescence staining
The expressive levels of microtubule-associated protein 1 light chain 3 (LC3) was measured by immunofluorescence staining (29). Fluorescence was measured under confocal microscopy (C1SiR; Nikon, Japan).
Western blot assay
Proteins were extracted from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). The protein concentration was tested by bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China), following the manufacturer’s instructions. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and then transferred to a polyvinylidene fluoride membrane (Bio-Rad). Membranes were blocked in 5% skim milk for 1 hour at room temperature, and then incubated with primary antibodies at 4 °C overnight. After incubating with horseradish peroxidase-conjugated antirabbit immunoglobulin G secondary antibody for 1 hour at room temperature. The proteins were measured with an Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare, UK).
Luciferase reporter assay
The 3'-UTR fragment of ATG5 containing the miR-153-3p binding sequence was cloned in a pMIR reporter vector, and a mutated plasmid was used as a control. The cells were then co-transfected with ATG5-WT or ATG5-MUT and miR-153-3p mimics using Lipofectamine 2000, according to the manufacturer's recommendation. Luciferase activity was tested using the Dual-Luciferase Reporter Assay System (Promega, Wisconsin, USA).
Statistical analysis
Data were presented as mean ± standard deviation, and analyzed using analysis of variance test and Student’s t-test in SPSS version 19.0 software. Values at P<0.05 were considered statistically significant.
Results
Upregulation of miR-153-3p sensitizes melanoma cells to dacarbazine
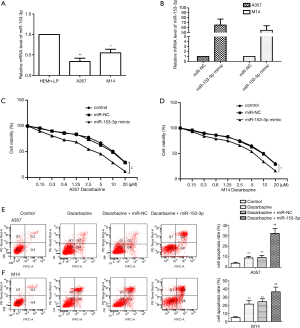
We initially examined the relative expression of miR-153-3p in HEMn-LP and melanoma cell lines (A357 and M14), respectively. The relative expression levels of miR-153-3p were obviously downregulated in A357 and M14 cells compared with HEMn-LP cells (P<0.05) (Figure 1A). The A357 and M14 cells were then transfected with miR-NC or miR-153-3p mimic, respectively, and the transfection efficiency was detected by qRT-PCR (P<0.05) (Figure 1B). The upregulation of miR-153-3p was found to decrease the half-maximal inhibitory concentration (IC50) values of dacarbazine in melanoma cells (Figure 1C,D). Furthermore, overexpressed miR-153-3p also notably increased cell apoptotic rates under dacarbazine treatment compared with the dacarbazine + miR-NC group (P<0.05) (Figure 1E,F). These results indicated that overexpressed miR-153-3p enhanced sensitivity to dacarbazine, inhibited cell proliferation, and increased apoptosis.
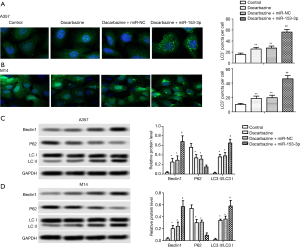
Upregulation of miR-153-3p enhanced dacarbazine-induced autophagy in melanoma cells
As shown in Figure 2, dacarbazine increased the number of LC3+ puncta per cell, Beclin1 expression levels, and the LC3 II/I rate, with decreased p62 expressive levels significantly compared with the control group. The upregulation of miR-153-3p was found to increase the number of LC3+ puncta per cell, Beclin1 expression levels, and the LC3 II/I rate, and decreased p62 expressive levels compared with the dacarbazine + miR-NC group, indicating that overexpressed miR-153-3p enhanced dacarbazine-induced autophagy in melanoma cells (P<0.05). These findings suggested that overexpressed miR-153-3p enhanced cell autophagy.
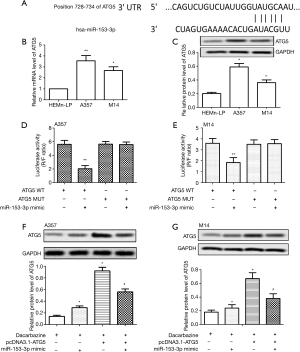
ATG5 is a target of miR-153-3p in melanoma cells
To detect the molecular mechanism underlying the miR-153-3p-regulated dacarbazine resistance in melanoma cells, putative miR-153-3p targets were predicted through bioinformatics analysis. ATG5 was found to be one of the potential targets of miR-153-3p (Figure 3A). Relative expression mRNA and the protein of ATG5 were significantly upregulated in A357 and M14 cells compared with HEMn-LP cells (P<0.05) (Figure 3B,C). Moreover, the Dual-Luciferase Reporter Assay showed that co-transfection with miR-153-3p mimic and ATG5-WT caused a significant downregulation in luciferase activity, whereas co-transfection with miR-153-3p mimic and ATG5-MUT did not influence luciferase activity significantly, confirming the targeting relationship between miR-153-3p and ATG5 in both A357 and M14 cells (P<0.01) (Figure 3D,E). When A357 and M14 cells were transfected with pcDNA3.1-ATG5, the expression of ATG5 increased with treatment with dacarbazine. When A357 and M14 cells were co-transfected with pcDNA3.1-ATG5 and miR-153-3p mimic, the expression of ATG5 decreased in the treatment with dacarbazine (P<0.05) (Figure 3F,G). These results suggested that miR-153-3p could negatively regulate the expression of ATG5.
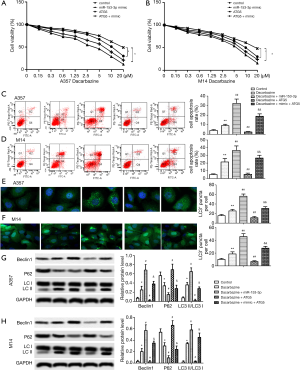
miR-153-3p increases sensitivity to dacarbazine in melanoma cells via the downregulation of ATG5
When A357 and M14 cells were transfected with pcDNA3.1-ATG5, cell viability increased in the treatment with dacarbazine compared with the control group. When A357 and M14 cells were co-transfected with pcDNA3.1-ATG5 and miR-153-3p mimic, cell viability decreased in the treatment with dacarbazine compared with the dacarbazine+ATG5 group (P<0.05) (Figure 4A,B). As shown in Figure 4C,D,E,F, when A357 and M14 cells were transfected with pcDNA3.1-ATG5, cell apoptosis and autophagy were downregulated in the treatment with dacarbazine compared with the control group. When A357 and M14 cells were co-transfected with pcDNA3.1-ATG5 and miR-153-3p mimic, cell apoptosis and autophagy were upregulated in the treatment with dacarbazine compared with the dacarbazine+ATG5 group (P<0.05) (Figure 4A,B).
Discussion
Melanoma is one of the most aggressive malignant skin tumors worldwide (30). Although surgical resection can increase the 5-year survival rates of patients with early-stage melanoma, there is no gold-standard treatment for advanced melanoma. Dacarbazine is a standard antineoplastic agent commonly used for melanoma treatment. However, the resistance of melanoma cells to dacarbazine inhibits the efficiency of this agent; therefore, it is important to investigate the underlying mechanism of dacarbazine resistance in melanoma cells. In the present study, we found that miR-153-3p overexpression sensitized melanoma cells to dacarbazine by suppressing ATG5-mediated apoptosis and autophagy, suggesting that targeting miR-153-3p may be a novel therapeutic strategy to treat melanoma patients.
miRNAs are involved in the post-transcriptional regulation of their target genes (31,32). Increasing evidence has demonstrated that various miRNAs are involved in the chemoresistance of many types of cancer cells, including melanoma cells. For example, miR-30a-5p confers cisplatin resistance by targeting insulin-like growth factor 1 receptor expression in melanoma cells (33). miR-579-3p ectopic expression was found to impair the establishment of drug resistance in human melanoma cells (34). Circular RNA 0084043 promotes malignant melanoma progression via the miR-153-3p/Snail axis (35). miR-153-3p was reported to affect tumor suppression, inhibiting cell proliferation and invasion by targeting SNAI1 in melanoma (22). However, whether miR-153-3p regulates dacarbazine resistance in melanoma cells has not been previously reported to the best of our knowledge, and requires further investigation. In the present study, the expression levels of miR-153-3p decreased in melanoma cells (A357 and M14), which was in agreement with Zeng et al.’s results that miR-153-3p expression levels decreased in melanoma tissues and melanoma cells (22). The upregulation of miR-153-3p decreased the IC50 value of dacarbazine, whereas it increased the cell apoptotic rate in both A357 and M14 cells, suggesting that the upregulation of miR-153-3p sensitized melanoma cells to dacarbazine.
20 It is well known that autophagy plays a regulatory role in the proliferation and death of cancer cells. Autophagy has also been linked to therapy resistance of cancer cells. Previous studies have demonstrated that chemotherapy might induce autophagy, which attenuates apoptosis, leading to drug resistance (36). Pei et al. reported that autophagy facilitates metadherin-induced fluorouracil resistance through the Adenosine monophosphate (AMP)-activated protein kinase (AMPK)/ATG5 pathway in gastric cancer (37). Autophagy is triggered during chemotherapy, and the inhibition of autophagy increases the chemosensitivity of castration-resistant prostate cancer cells (38). Besides LC3, Beclin1 is also important for the formation of autophagosomes (39). In the present study, dacarbazine treatment induced autophagy in melanoma cells by increasing autophagosomes, Beclin1 expression, and LC3 I to LC3 II conversion, and decreasing P62 expression. Moreover, miR-153-3p overexpression was observed to enhance dacarbazine-induced autophagy in melanoma cells. These results demonstrate that miR-153-3p overexpression enhances the sensitivity of melanoma cells to dacarbazine.
Previous studies have reported that ATGs play vital roles during autophagy. ATG5 is a key player in autophagosome formation (40). One study indicated that ATG5 promotes Wnt signaling and autophagy in melanoma (41). In our bioinformatics study, we revealed that ATG5 is a potential target of miR-153-3p. There is a negative relationship between ATG5 and miR-153-3p expression in melanoma cells. Furthermore, the overexpression of ATG5 decreases dacarbazine sensitivity and promotes proliferation, and inhibits apoptosis and autophagy in melanoma cells. miR-153-3p exhibits suppressive effects via directly binding and downregulating ATG5 expression, which subsequently increases sensitivity to dacarbazine and inhibits proliferation, and enhances apoptosis and autophagy in melanoma cells.
Conclusions
Taken together, there results of the present study show that the miR-153-3p/ATG5 axis plays a vital role in the regulation of dacarbazine resistance in melanoma cells. miR-153-3p was found to exhibit suppressive effects via directly binding and downregulating ATG5 expression, which subsequently increased sensitivity to dacarbazine and inhibited proliferation, as well as enhancing apoptosis and autophagy in melanoma cells, providing a basis to explore the functions of miRNAs on drug resistance in the treatment of melanoma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2660
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2660
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2660). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was obtained from the Ethics Committee of Medical College of Shanxi Datong University (No. 20190123).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kosary CL, Altekruse SF, Ruhl J, et al. Clinical and prognostic factors for melanoma of the skin using SEER registries: collaborative stage data collection system, version 1 and version 2. Cancer 2014;120:3807-14. [Crossref] [PubMed]
- Coit DG, Thompson JA, Andtbacka R, et al. Melanoma, version 4.2014. J Natl Compr Canc Netw 2014;12:621-9. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Xu TX, Yu SF, Ma M, et al. cMET-N375S germline mutation is associated with poor prognosis of melanoma in Chinese patients. Transl Cancer Res 2018;7:248-56. [Crossref]
- Song X, Zhao Z, Barber B, et al. Overall survival in patients with metastatic melanoma. Curr Med Res Opin 2015;31:987-91. [Crossref] [PubMed]
- Eriksson H, Frohm-Nilsson M, Jaras J, et al. Prognostic factors in localized invasive primary cutaneous malignant melanoma: results of a large population-based study. Br J Dermatol 2015;172:175-86. [Crossref] [PubMed]
- Pan Y, Haydon AM, McLean CA, et al. Prognosis associated with cutaneous melanoma metastases. Australas J Dermatol 2015;56:25-8. [Crossref] [PubMed]
- Yentz S, Lao CD. Immunotherapy for mucosal melanoma. Ann Transl Med 2019;7:S118. [Crossref] [PubMed]
- Friedlander P. The use of baseline tumor size to prognosticate overall survival in stage IV melanoma patients treated with the PD-1 inhibitor pembrolizumab. Ann Transl Med 2019;7:S24. [Crossref] [PubMed]
- Eigentler TK, Caroli UM, Radny P, et al. Palliative therapy of disseminated malignant melanoma: a systematic review of 41 randomised clinical trials. Lancet Oncol 2003;4:748-59. [Crossref] [PubMed]
- Serrone L, Zeuli M, Sega FM, et al. Dacarbazine-based chemotherapy for metastatic melanoma: thirty-year experience overview. J Exp Clin Cancer Res 2000;19:21-34. [PubMed]
- Li H, Chen J, Wang X, et al. Nodal induced by hypoxia exposure contributes to dacarbazine resistance and the maintenance of stemness in melanoma cancer stem-like cells. Oncol Rep 2018;39:2855-64. [Crossref] [PubMed]
- Erdmann S, Seidel D, Jahnke HG, et al. Induced cross-resistance of BRAF(V600E) melanoma cells to standard chemotherapeutic dacarbazine after chronic PLX4032 treatment. Sci Rep 2019;9:30. [Crossref] [PubMed]
- Chen YN. Dacarbazine inhibits proliferation of melanoma FEMX-1 cells by up-regulating expression of miRNA-200. Eur Rev Med Pharmacol Sci 2017;21:1191-7. [PubMed]
- Eberle J, Kurbanov BM, Hossini AM, et al. Overcoming apoptosis deficiency of melanoma-hope for new therapeutic approaches. Drug Resist Updat 2007;10:218-34. [Crossref] [PubMed]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010;22:124-31. [Crossref] [PubMed]
- Gao Y, Qi W, Liu S, et al. Acid-induced autophagy protects human gastric cancer cells from apoptosis by activating Erk1/2 pathway. Transl Cancer Res 2019;8:1560-70. [Crossref]
- Ding X, Yue W, Chen H. Effect of artesunate on apoptosis and autophagy in tamoxifen resistant breast cancer cells (TAM-R). Transl Cancer Res 2019;8:1863-72. [Crossref]
- Vera-Ramirez L, Vodnala SK, Nini R, et al. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9:1944. [Crossref] [PubMed]
- Brahmbhatt H, Oppermann S, Osterlund EJ, et al. Molecular Pathways: Leveraging the BCL-2 Interactome to Kill Cancer Cells--Mitochondrial Outer Membrane Permeabilization and Beyond. Clin Cancer Res 2015;21:2671-6. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Zeng HF, Yan S, Wu SF. MicroRNA-153-3p suppress cell proliferation and invasion by targeting SNAI1 in melanoma. Biochem Biophys Res Commun 2017;487:140-5. [Crossref] [PubMed]
- Wang XB, Wang H, Long HQ, et al. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J Cell Physiol 2019;234:7115-27. [Crossref] [PubMed]
- Shen F, Chang H, Gao G, et al. Long noncoding RNA FOXD2-AS1 promotes glioma malignancy and tumorigenesis via targeting miR-185-5p/CCND2 axis. J Cell Biochem 2019;120:9324-36. [Crossref] [PubMed]
- Sun D, Wang X, Sui G, et al. Downregulation of miR-374b-5p promotes chemotherapeutic resistance in pancreatic cancer by upregulating multiple anti-apoptotic proteins. Int J Oncol 2018;52:1491-503. [Crossref] [PubMed]
- Zheng Y, Sun Y, Liu Y, et al. The miR-31-SOX10 axis regulates tumor growth and chemotherapy resistance of melanoma via PI3K/AKT pathway. Biochem Biophys Res Commun 2018;503:2451-8. [Crossref] [PubMed]
- Lv T, Song K, Zhang L, et al. miRNA-34a decreases ovarian cancer cell proliferation and chemoresistance by targeting HDAC1. Biochem Cell Biol 2018;96:663-71. [Crossref] [PubMed]
- Piotrowska A, Wierzbicka J, Rybarczyk A, et al. Vitamin D and its low calcemic analogs modulate the anticancer properties of cisplatin and dacarbazine in the human melanoma A375 cell line. Int J Oncol 2019;54:1481-95. [Crossref] [PubMed]
- Tong H, Zhu Y, Liu Y. Incidence and risk of fatigue in cancer patients treated with MET inhibitors: A systematic review and meta-analysis. Medicine (Baltimore) 2019;98:e15522. [Crossref] [PubMed]
- Naves LB, Dhand C, Venugopal JR, et al. Nanotechnology for the treatment of melanoma skin cancer. Prog Biomater 2017;6:13-26. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Li Y, Zhang J, Liu Y, et al. MiR-30a-5p confers cisplatin resistance by regulating IGF1R expression in melanoma cells. BMC Cancer 2018;18:404. [Crossref] [PubMed]
- Fattore L, Mancini R, Acunzo M, et al. miR-579-3p controls melanoma progression and resistance to target therapy. Proc Natl Acad Sci U S A 2016;113:E5005-13. [Crossref] [PubMed]
- Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun 2018;502:22-9. [Crossref] [PubMed]
- Zeng Y, Yang X, Wang J, et al. Aristolochic acid I induced autophagy extenuates cell apoptosis via ERK 1/2 pathway in renal tubular epithelial cells. PLoS One 2012;7:e30312. [Crossref] [PubMed]
- Pei G, Luo M, Ni X, et al. Autophagy Facilitates Metadherin-Induced Chemotherapy Resistance Through the AMPK/ATG5 Pathway in Gastric Cancer. Cell Physiol Biochem 2018;46:847-59. [Crossref] [PubMed]
- Hu F, Zhao Y, Yu Y, et al. Docetaxel-mediated autophagy promotes chemoresistance in castration-resistant prostate cancer cells by inhibiting STAT3. Cancer Lett 2018;416:24-30. [Crossref] [PubMed]
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1-222.
- Cemma M, Brumell JH. Interactions of pathogenic bacteria with autophagy systems. Curr Biol 2012;22:R540-5. [Crossref] [PubMed]
- Ndoye A, Budina-Kolomets A, Kugel CH 3rd, et al. ATG5 Mediates a Positive Feedback Loop between Wnt Signaling and Autophagy in Melanoma. Cancer Res 2017;77:5873-85. [Crossref] [PubMed]
(English Language Editor: R. Scott)



