Prognostic value of let-7 in lung cancer: systematic review and meta-analysis
Introduction
Lung cancer ranks first among malignant tumors in terms of mortality rate. Early detection and accurate prognosis analysis are key to improving the survival rate of patients with lung cancer (1). Lung cancer is one of the most malignant tumors with the fastest growth in morbidity and mortality, and poses the greatest threat to public health and people’s life beings. Lung adenocarcinoma is the most concentrated type of lung cancer, with carcinogenic driven mutations. Therefore, related targeted mutation gene diagnosis, drug development and clinical use are also most effective.
The rapid development of sequencing technology and the reduction of cost enables us to quickly accumulate more clinical genetic mutation diagnostic data. The collection and analysis of more diagnosis data and corresponding clinical practice will further guide us to treat lung cancer reasonably, and conduct effective diagnosis and treatment for more patients, which has gradually become a feasible trend. For example, with the continuous promotion of next-generation sequencing, including the beginning of universal application of liquid biopsy, we have found an increasing number of double-targeted mutations or multi-targeted mutations and new biomarkers related to prognosis factors for patients with lung cancer.
MicroRNAs (miRNAs) is a class of endogenous small RNAs that play a great many essential regulatory roles in cells. It is speculated that miRNA regulates one-third of human genes. The high degree of conservation of miRNAs is closely related to the importance of their functions. miRNAs are closely related to the evolution of their target genes and then affecting the growth and development of organisms at the level of cells, tissues or individuals, and participates in the occurrence and development of a variety of diseases including tumors (2,3). Studies have shown that by introducing let-7 into lung cancer cells to change their low expression level, tumor cell growth can be significantly inhibited (4). However, we found that there is little research on the relationship between let-7 expression and the prognosis of patients with lung cancer. At the same time, we put forward our hypothesis of whether the expression level of let-7 is significantly correlated with the prognosis of patients with lung cancer and whether the low expression of let-7 in lung cancer suggests poor prognosis (4,5). Consequently, we conducted this meta-analysis to evaluate the value of let-7 in the prognosis of patients with lung cancer. We present the following article in accordance with the PRISMA reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1240).
Methods
Searching strategies
We searched the PubMed, Web of Science, EMBASE and CNKI databases and studies published from Jun 1993 to July 2019, with entries of (I) “let-7 or MiRNA let-7”; (II) “Lung Cancer or Pulmonary Neoplasms”; (III) “Prognoses or Prognostic Factors”. In addition, as we collect the research data for this paper, in order to avoid the case that two checkers found the same article because of the data in different publications, we further checked these articles in details, and finally ensured no repeated studies. Literatures were searched independently by two researchers, and consensus was reached through discussion when there was disagreement.
Inclusion and exclusion criteria
Inclusion criteria: (I) lung cancer is the main target of this project; (II) the specific details of demographic data or the survival curve can be obtained and counted in the original research results; (III) the overall follow-up period of the study was more than 12 months; (IV) all the included research articles can be viewed in full text.
Exclusion criteria: (I) the article types of reviews, case reports, letters, and conference reports are excluded; (II) researches not based on humans; (III) lacks original data or graphs that can calculate the hazard ratio (HR), 95% confidence interval (95% CI) and P value; (IV) basic laboratory research is excluded.
Quality assessment
The Newcastle-Ottawa Scale (NOS) Guide is designed to evaluate such research systems. The evaluation tool is mainly used to assess the quality of the research by using star coefficients, with 9 stars representing the highest score. Specifically, high-quality research can be scored 8–9 stars; eligible research 6–7 stars, while 6-star or less indicates poor quality (Table 1).
Table 1
| Author (year) | Country | Design | Sample | Ethnicity | Method | Age (years) | Gender (M/F) | Cancer type | NOS |
|---|---|---|---|---|---|---|---|---|---|
| Jusufović (2012) | Bosnia and Herzegovina | ROS | 50 | European | RT-PCR | 63 | 50(10/40) | NSCLC | 8 |
| Landi (2010) | America | ROS | 290 | American | RT-PCR | – | 290(211/79) | NSCLC | 9 |
| Shin (2016) | Korea | ROS | 761 | Asian | RT-PCR | 65 | 751(203/551) | NSCLC | 8 |
| Takamizawa (2004) | Japan | ROS | 143 | Asian | RT-PCR | 62 | 143(90/53) | NSCLC | 9 |
| Voortman (2010) | America | ROS | 783 | France | RT-PCR | – | – | NSCLC | 7 |
| Yanaihara (2006) | America | ROS | 65 | American | RT-PCR | 67 | – | Adenocarcinoma | 8 |
| Zhao (2014) | China | ROS | 94 | Asian | RT-PCR | 94 | 94 (65/29) | NSCLC | 7 |
ROS, review observational study; NSCLC, non-small cell lung cancer; NOS, Newcastle-Ottawa Scale.
Data collection
Any matters that were unclear or disagreement were dealt with by means of discussion. Excel was used to collect the following details (Table 1): authors, the year of publication, nations, study design, study period, detecting methods, follow-ups, enrolled samples, age, gender, the tumor stage, tumor histology, the number of patients with positive expression and negative expression of let-7; treatment information, and survival analyses.
Statistical analysis
Meta-analysis was used to merge the results of the included literatures and draw the forest map. The heterogeneity of effect size distribution among studies was measured by Q test. I2 statistics were used to evaluate the percentage for inter-study variation in the total variation: if P>0.05, I2<50%, it indicates there is no statistical heterogeneity among the studies, the fixed effect model should be used for data consolidation, if P<0.05, I2>50% or higher, it explains that each study has statistical heterogeneity, and further analysis should be done to figure out the reasons of the heterogeneity of statistics. If the results of the study are in the same direction, then there is no obvious clinical heterogeneity and negligible statistical heterogeneity by using the fixed effects model. Instead, if the direction of the research results were opposite, then a random effects model should be applied to merge and carry out meta-analysis. Funnel plot was used to determine whether publication bias existed in the included literatures, and when the funnel plot showed asymmetry or incompleteness, publication bias was considered to exist. All P values were tested bilaterally. Stata 12.0 statistical software was used for the statistical analysis. HR and 95% CI were used as the effect quantity to evaluate the correlation between let-7 low expression and lung cancer prognosis with HR >1 indicating the poor prognosis.
Results
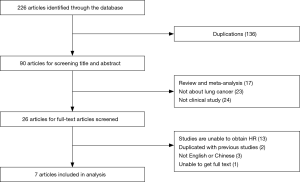
The 226 researches had been selected from four online databases including PubMed, EMBASE, Web of Science and CNKI. In the first step we picked out 90 articles from 226 articles by screening titles and abstracts. In the second step, based on our inclusion criteria, 64 papers out of 90 articles were excluded due to unqualified part of the document type, and the remaining documents were fully read in full text. In the third step, 26 potential qualified papers were identified in our meta-analysis. Finally, only seven articles are in line with all of our requirements (4,6-11) (Figure 1).
The characteristics of included studies
All the basic features of the seven qualified documents are listed in Table 1. Takamizawa et al. (4) reported the clinical and biological effects of let-7 in lung cancer tissues and the potential clinical and biological effects of this miRNA for the first time. Jusufović et al. (6) estimated the lower let-7b expression with worse prognosis in Serbia and Bosnia and Herzegovina lung cancer patients. Landi et al. (7) identified a let-7 family expression profile that strongly differentiated adenocarcinoma from squamous cell carcinoma and had prognostic implications. Shin et al. (8) enrolled 761 patients with surgically estimated that let-7 have a prognostic impact on surgically resected NSCLC in Asia. Voortman et al. (9) enrolled 783 patients with non-small cell lung cancer who underwent lobectomy and adjuvant chemotherapy, let-7 was not a meaningful predictor of prognosis. Yanaihara et al. (10) indicates that let-7 expression profiles are diagnostic and prognostic markers of adenocarcinomas. Zhao et al. (11) has reached the same conclusion that let-7 reduction predicted a poor prognosis for lung cancer patients after surgery in China.
Meta-analysis results
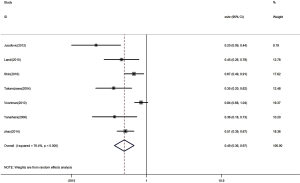
The HR for the seven appropriate studies was 0.49 (95% CI: 0.36–0.67, P<0.00001), suggesting that low expression of let-7 in lung cancer is a predictor of low survival rate in patients with NSCLC (Figure 2). Heterogeneity of overall prognosis was relatively low (I2=76.4%, P=0.000). We conducted further grouping surveys on these collected data.
Let-7 expression and disease-free survival (DFS) in patients with lung cancer
Three studies were conducted on DFS of the included samples, taking I2=90% into consideration, by using a random-effect model and combining statistics. It was found that the low-expression let-7 level predicted a worse DFS (HR =0.41, 95% CI: 0.21–0.78) (Figure 3).
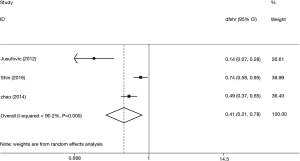
Let-7 expression and ethnicity
We measured the association between let-7 expression and ethnicity. The samples in our research can be divided into Europe, America and Asia, while the summarized HR of analysis was 0.61 (95% CI: 0.53–0.70) (Figure 4). When we compared the relationship between let-7 expression and different countries, it was statistically significant in Europe (HR =0.76, 95% CI: 0.62–0.93), America (HR =0.41, 95% CI: 0.27–0.64) and Asia (HR =0.54, 96% CI: 0.45–0.66). These results showed that lower let-7 expression was a sign of poor survival.
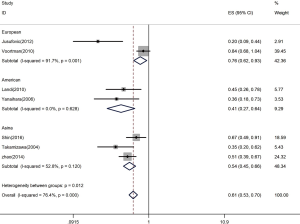
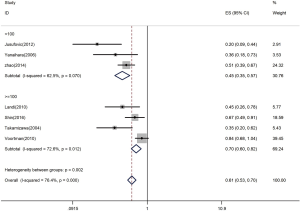
Let-7 expression and quantity of samples in studies
Firstly, the HR was 0.45 with samples less than 100 (95% CI: 0.35–0.57) (Figure 5). With samples exceeding or equal to 100, the HR-based analysis also revealed the significantly lower let-7 expression in patients was an indicator of poor survival (HR =0.70, 95% CI: 0.60–0.82) (Figure 5).
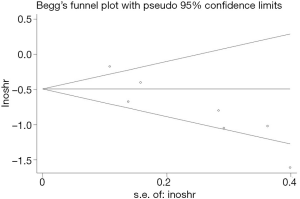
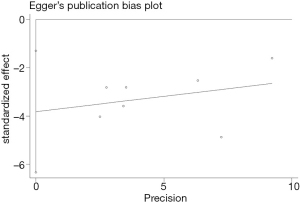
Publication bias
In this study, two methods of assessing publication bias were completed (Begg’s funnel plot and Egger’s test) (Figures 6,7). The Begg’s test scores for the seven research papers were P=0.072, and the Egger’s test scores were P=0.011. According to Begg’s test results, the significance of publication bias was not detected in overall survival. Nonparametric pruning and filling methods were used to assess this bias and summarize the impact of HR on overall survival. Three of the seven articles were about DFS, with an egger’s score (P=0.221) and a Begg’s score (P=0.296). In summary, the statistical results of this study were statistically reliable from the perspective of overall survival.
Discussion
In conclusion, lung cancer is the most common malignant tumor with the highest morbidity and mortality rate in the world. Therefore, early detection and effective treatment of lung cancer is key to improving the prognosis and the survival rate. The let-7 family is an early-discovered miRNA that can detect its expression in adult tissues, and some of the let-7 exhibits a low expression level in tumor cells or tissues, but in some tumors, the expression of let-7 is up-regulated. This meta-analysis included 7 eligible articles and was the first to evaluate the prognostic value of let-7 in NSCLC patients. Let-7 is one of the miRNAs closely related to lung cancer. The results of this study suggest that the low expression of let-7 is statistically correlated with some clinical outcomes, and that the low expression of let-7 is a risk factor for lung cancer prognosis, suggesting poor prognosis. However, the mechanism of let-7 family is complex, let-7, as a tumor suppressor, negatively regulates multiple oncogenes, such as RAS and high mobility group protein A2 (HMGA2). It also negatively regulates a variety of cell cycle regulators, such as CDC25A, CDK6, cyclin D2 (12-14). It plays a vital role in lung cancer tissue. A retrospective study (5) showed that low expression of let-7c is related to metastasis, vascular invasion and progression of lung cancer, and is an independent factor for poor prognosis of lung cancer patients. In Voortman’s study (9) included in the literature, in some large cohort studies of NSCLC patients, miRNA expression patterns are neither predictive nor prognostic. On one hand, the reason may be related to the effect of radiotherapy to the patient’s body, on the other hand, it may be related to the later stage of tumor. Studies have shown that let-7 performances as a tumor suppressor in the lung tissue, and as an independent prognostic factor of lung cancer, let-7 needs to be confirmed by more randomized controlled trials.
There are still some shortcomings in this meta-analysis that requires readers’ attention. First, the number included in this study is small, and the sample scale is smaller than other articles. Second, there is irremovable heterogeneity between the various literatures. Consideration may be given to the differences in patients’ clinical characteristics (such as age, the sex ratio, the tumor stage, pathological type and ethnicity, etc.), data analysis, technical platform, sensitivity and specificity of cut-off points and detection. Third, the number of papers included to explore the relationship between let-7 expression, the patient gender and lung cancer differentiation was small, which may affect the authenticity of the results. Fourth, publication bias exists among studies, which may affect the final results. The most likely reason for publication bias is that reports with positive results receive more attention, while negative results are rarely published or cited and are prone to be leaved out. In the future, experimental design and the characteristics of the subjects should be described in more details, the diagnostic criteria and treatment range should be clarified, the sample content should be increased, and the bias in each stage of the experiment should be controlled.
Conclusions
In conclusion, the preliminary study of let-7 in lung cancer showed that its high expression and the cell function will be inhibited, resulting in increased expression of oncogene RAS, thereby increasing the activity of lung cancer cells, shortening the survival period of lung cancer patients, and worsening the prognosis. All of this suggests that let-7 is closely related to lung cancer. However, there are still some questions worth further study: What are the upstream signaling molecules and mechanisms of action that cause let-7 function inhibition in lung cancer cells? Does let-7 still have other targets? Therefore, with in-depth research on let-7, we have seen clearer significance of let-7 in the prognosis of lung cancer, and more literature is needed to make the research results more persuasive.
Acknowledgments
We greatly appreciate the assistance of the staff of the Department of Thoracic Surgery, West China Hospital, Sichuan University for their efforts.
Funding:
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1240
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1240). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A 2007;104:11400-5. [Crossref] [PubMed]
- Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol 2013;48:51-68. [Crossref] [PubMed]
- Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer 2010;103:1144-8. [Crossref] [PubMed]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [Crossref] [PubMed]
- Zhu WY, Luo B, An JY, et al. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest 2014;32:394-401. [Crossref] [PubMed]
- Jusufović E, Rijavec M, Keser D, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non--small--cell lung cancer. PLoS One 2012;7:e45577. [Crossref] [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [Crossref] [PubMed]
- Shin KM, Jung DK, Hong MJ, et al. The pri-let-7a-2 rs1143770C>T is associated with prognosis of surgically resected non-small cell lung cancer. Gene 2016;577:148-52. [Crossref] [PubMed]
- Voortman J, Goto A, Mendiboure J, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res 2010;70:8288-98. [Crossref] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [Crossref] [PubMed]
- Zhao B, Han H, Chen J, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett 2014;342:43-51. [Crossref] [PubMed]
- Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A 2008;105:3903-8. [Crossref] [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [Crossref] [PubMed]
- Sarhadi VK, Wikman H, Salmenkivi K, et al. Increased expression of high mobility group A proteins in lung cancer. J Pathol 2006;209:206-12. [Crossref] [PubMed]