Identification and validation of potential novel prognostic biomarkers for patients with glioma based on a gene co-expression network
Introduction
Glioma, whose prevalence varies substantially with patient age, gender, race, and a number of other clinicopathological parameters, is widely regarded as one of most lethal and challenging diseases of the nervous system; the median survival time of glioma patients is 5–10 years, and the estimated 10-year survival rate varies between 5% and 50% (1,2). Moreover, following a tumorectomy, the combined use of both concurrent radiation and temozolomide treatment is currently the standard therapy for patients with glioma. Notably, even though gliomas are generally subdivided into two subgroups based on a known profile of related biomarkers (3), i.e., low-grade gliomas and high-grade gliomas, patients with the same diagnostic results sometimes have totally different outcomes even after the same therapeutic procedures. For instance, after approximately 6.9 months, patients with high-grade gliomas are more likely to have recurrent glioma, resulting in a median survival time of about 12–15 months in these patients following diagnosis (2). Therefore, despite surgical success, the clinical outcome of glioma patients is generally unsatisfactory (4).
Many studies have tried to find a therapeutic target or prognostic biomarker for glioma to improve clinical outcomes (5,6). Mutations in the genes isocitrate dehydrogenase (IDH) 1 and 2 have been reported in 73–85% of secondary glioblastomas and 72–100% of stage II and III astrocytic and oligodendroglial tumors, but these mutations are almost never found in primary glioblastomas (5,6). IDH mutations represent a major biomarker with diagnostic, prognostic, and predictive implications (6). In addition, promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT) is found in about 80% of low-grade IDH-mutated gliomas. Low MGMT levels correlate with modestly improved survival (5). Moreover, the epidermal growth factor receptor (EGFR) is amplified in about 40% of glioblastoma patients, and it is often associated with high-grade classical tumors. However, whether EGFR amplification can be used as a prognostic biomarker is controversial (5,6). Recently, the Rho-specific guanine-nucleotide exchange factor PLEKHG5 was reported to be involved in glioma migration, invasion, and epithelial-mesenchymal transition and could be used as a prognostic biomarker for glioma patients (7). Furthermore, some other signaling pathways including the p53 pathway, phosphoinositide 3-kinase pathway, retinoblastoma pathway, and Ras/Raf/mitogen-activated protein kinase (MAPK) pathway also have been reported to be involved in the progression of glioma (5,6). However, these studies may have overemphasized the determination of a differential expression profile while ignoring the crosstalk between genes and/or signaling pathways, which are likely to be functionally correlated. Therefore, the clinical application of these biomarkers is limited.
During the last decade, studies have started to focus on unscrambling related gene expression profiles, leading to a better understanding of the potential molecular mechanisms underlying the formation and progression of glioma. By using weighted gene co-expression network analysis (WGCNA) based on The Cancer Genome Atlas (TCGA) database, Xu et al. have found that gene prognosis models with four genes (OSMR + SOX21 + MED10 + PTPRN) can be used for estimating overall survival in glioblastoma patients (8). However, since samples in the TCGA database are mainly from the USA, some conclusions drawn from the TCGA database may not be suitable for the Chinese population. Therefore, we conducted a similar study using the Chinese Glioma Genome Atlas (CGGA) database, which is based on a Chinese population.
Accordingly, in this study, WGCNA was used to determine the association between functional gene clusters and clinicopathological parameters through the network construction of co-expressing genes based on the similarities in gene expression as well as analogous functions among all of the genes from collected samples of glioma patients, revealing the responsible genes within the most significant module in the network. Moreover, functional enrichment analysis of these identified genes also was carried out within the co-expression networks, thereby uncovering the underlying molecular mechanisms for the formation and progression of gliomas. Thus, the results of this study will provide a better understanding of the pathogenesis of gliomas and might offer a novel clinical treatment for Chinese glioma patients. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-492).
Methods
Data collection and pre-processing
In accordance with the 2007 World Health Organization classification, the clinicopathological parameters of the eligible patients with gliomas along with their expression profiles (233 cases) were extracted from the CGGA database (http://www.cgga.org.cn). The following clinicopathological parameters of patients with histologically confirmed glioma, who had undergone a tumorectomy as well as follow-up treatment at Beijing Tiantan Hospital who established the CGGA database, were collected: age, gender, pathologic diagnosis, and outcome. Among them, the complete data of 53 patients (Table S1) who received chemotherapy and radiotherapy were selected for WGCNA to identify hub genes, which were further validated using the TCGA database (https://genome-cancer.ucsc.edu/) (9). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the review board of the local institution and the CGGA database of the Beijing Neurosurgical Institute at Capital Medical University. However, the requirement of written informed consent from the included patients with gliomas was waived.
Identification of clinically significant modules through WGCNA
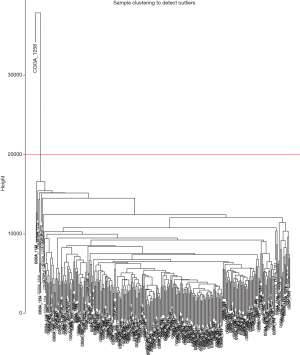
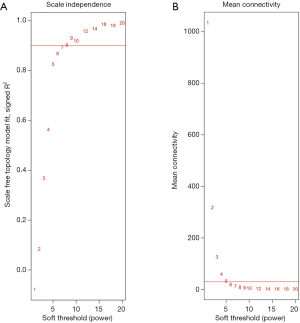
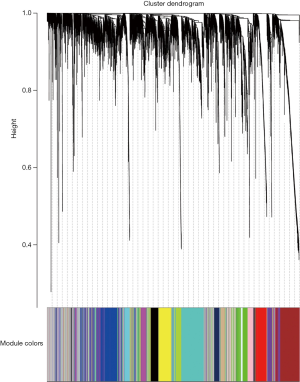
Following data pre-processing of the high-throughput mRNA sequencing data, the genes with increased genetic variance were collected for the systematic network construction of gene co-expression (Figure 1) by using the WGCNA (version 1.41; labs.genetics.ucla.edu/horvath/coexpressionNetwork/Rpackages) algorithm (10). First, a scale-free topology was constructed for the network analysis with a proper β power (the power of β=9) (Figure 2). Second, based on the topology overlap, hierarchical average linkage clustering was utilized to identify modules of interest, thus obtaining the specific gene modules associated with certain phenotypes, followed by visualization with plots within the gene network. Finally, the association between the modules and the overall survival of the glioma patients receiving a combination of radiotherapy and chemotherapy was determined.
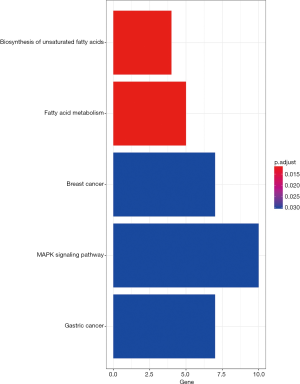
After the gene modules were identified, module significance was considered as the average gene significance among selected genes within one specific module (Figure 3). The module with the highest value of module significance compared to the other selected modules was regarded as a potential candidate that might be tightly associated with clinicopathological parameters (Figure 4). The gene module that was tightly associated with the overall survival of the glioma patients was then chosen for additional analysis. To further determine the potential signaling pathways of genes shown in the modules to be related with the clinicopathological parameters (Figure 5), the “clusterProfiler” package (11) in R was used to annotate and visualize the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (12).

Hub gene identification
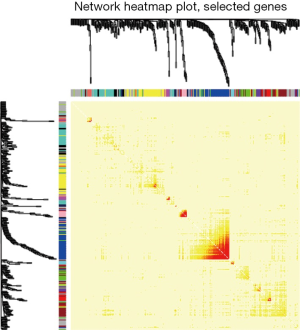
Hub genes, known as being extensively interconnected with other genes within the selected module, normally play essential roles in the network. Accordingly, modules of interest were chosen, and, in turn, critical hub genes were well defined by the connectivity of these modules based on the assessment of Pearson’s correlation coefficients. Briefly, coefficients for evaluation of the association between module connectivity and hub genes should be greater than 0.8, while coefficients for evaluation of the association between clinicopathological parameters and hub genes should be greater than 0.2. Moreover, the STRING database was also utilized for the construction of a model for protein-protein interactions (PPIs) when the confidence index of the related interaction was greater than 0.4. Through the construction of PPI networks, 50 defined hub genes were subsequently ranked by using minimum multicollinearity methods (Figure 6). The common hub genes detected in either the co-expression network or the PPI network were considered as authentic hub genes for further validation.
Evaluation of clinical efficacy
The hub genes were first validated using the survival data of all glioma patients from the CGGA database and then further validated using the survival data of low-grade glioma patients from the TCGA database, especially for the assessment of overall survival rates to determine the effect of all hub genes on the prognosis of patients with glioma. The Oncomine database (http://www.oncomine.org) was also utilized for further validation of candidate gene expression (13).
Statistical analysis
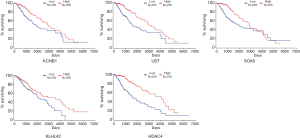
Regarding the overall survival rate, patients with a complete survival time and a censor status were subdivided into two groups in accordance with the median expression level of each hub gene, namely, high group vs. low group (Figure 7). R software was used for the log-rank test as well as Kaplan-Meier survival analysis (14).
Results
Identification of essential modules through WGCNA
Following WGCNA of the mRNA co-expression networks, 18 modules were identified and labeled with distinct colors from the mRNA network. Among them, the black module, showing the most significant correlation with the clinical prognosis, was regarded as being the best prognostic indicator for tumor prognosis by using the correlation analysis for the sequencing results of 233 gliomas (Figures 3,4). Through computation with the KEGG pathway database, functional enrichment analysis further revealed quite a few signaling pathways, such as the MAPK signaling pathway, underlying not only the formation and progression of breast/gastric cancer but also many vital biological processes, such as the biosynthesis of unsaturated fatty acids as well as fatty acid metabolism, suggesting that these correlated signaling pathways and biological processes potentially play indispensable roles in the formation/progression of gliomas (Figure 5).
Identification and validation of hub genes
The 29 genes with the most extensive interconnections within the black module were defined as hub genes by the connectivity profile along with their association with the clinicopathological parameters, based on Pearson’s correlation coefficient assessment (Figure 4). Besides, in accordance with the STRING database (https://string-db.org/), the PPI network was also constructed for further determination of all genes within the black module via Cytoscape (Figure 6). Thus, 50 defined hub genes were subsequently ranked by using minimum multicollinearity methods. Among them, five common hub genes (KCNB1, UST, SOX8, KLHL42, and HDAC4) that were found by analysis of either the co-expression or the PPI network were then accepted as authentic hub genes for further validation (15).
Evaluation of clinical efficacy and survival analysis
To further determine the correlation between the five hub genes and the related prognosis of glioma patients, survival curve analysis was performed by using Kaplan–Meier analysis. Prognostic cut-off criteria were set as the median values of the expression level of each hub gene. Through survival curve analysis, our results uncovered a significant difference in the overall survival rates of the glioma patients with distinct expression profiles of the selected hub genes, i.e., KCNB1, UST, SOX8, KLHL42, and HDAC4 (Figure 7), indicating that all these hub genes were potential prognostic biomarkers of glioma. Moreover, the Kaplan–Meier survival curve showed that a better discernibility was observed in data from the CGGA database (Figure 7).
Discussion
Glioma is known as the most lethal primary brain tumor, and glioma patients usually have a poor survival rate. In general, there are a variety of genetic alterations involved in glioma formation that primarily contribute to different responses to therapies. In this study, we aimed to identify novel biomarkers that offer better prognosis prediction in patients with glioma through analysis of a gene co-expression network. We hypothesized that the molecular mechanisms underlying any biological process require a close association of a co-expressed gene module sharing the same functional annotations. In this study, following comprehensive screening, 29 hub genes were specifically selected from the identified black module. Regarding the combined use of a co-expression network as well as a PPI network for further validation, five of them were considered as authentic hub genes (KCNB1, UST, SOX8, KLHL42, and HDAC4), suggesting that they are potentially promising indicators for assessment of vital biological processes as well as clinical outcomes. Furthermore, these five genes possessed a better prediction accuracy in Chinese patient samples from the CGGA database compared to samples from the TCGA database.
Specifically, our results showed that there was an apparent correlation of the identified black module with IDH mutations, which are widely accepted as being a distinct proneural genetic signature manifesting the glioma-CpG island methylator phenotype. Previous reports have revealed that IDH is not only associated with glioma formation/progression but also with its recurrence and secondary glioblastomas (16); hence, it is a potential novel therapeutic target for the treatment of IDH-mutant gliomas (17).
On the other hand, accumulating evidence has revealed the involvement of some of these identified hub genes in various vital processes. For instance, there was an apparently enhanced amount of KCNB1 expression along with accumulated autophagic vacuoles during the early and late phases of apoptosis, possibly through an ERK/MAPK-dependent signaling pathway (9). In addition, a heterogeneous pattern of Sox expression is generally regarded to be compatible with less-differentiated gliomas, as compared to their counterparts from the control group (18).
Moreover, the involvement of an epigenetic modification, such as histone modification, has been extensively investigated in tumorigenesis and chemotherapy/radiotherapy resistance for more than a decade. Previous studies using immunohistochemical analysis have uncovered that an increased expression level of HDAC4 predicts a poor overall survival for patients with glioma. However, HDAC4 is a highly abundant gene with a variety of vital biological functions in the brain. For example, HDAC4 is critical in the modulation of neuronal survival, thus strongly indicating that it at least partially contributes to therapeutic resistance and poor clinical outcomes in glioblastoma patients. Interestingly, significantly decreased expression of the HDAC gene has been reported to be closely associated with the etiology as well as tumor progression in grade-related astrocytoma (19). Likewise, the expression level of HDAC4 in combination with the methylation status of the promotor of the O6-methylguanine-DNA methyltransferase gene has been applied for the evaluation of glioblastoma for the prediction of its response to combined radiochemotherapy and prognosis after the therapy (20). Accordingly, there also have been a few reports on the essential role of HDAC4 mediating DNA-damage repair, which in turn reverses irradiation-induced DNA damage as well as stemness, resulting in the promotion of radioresistance in patients with glioblastoma (21,22).
Although we used a similar method as Xu et al. (8), the five genes (KCNB1, UST, SOX8, KLHL42, and HDAC4) we identified were totally different from the four genes (OSMR, SOX21, MED10, and PTPRN) reported by Xu et al. (8). The difference in the data source was the main reason for the discrepancy. Furthermore, during gene identification, we focused on patients who received chemotherapy and radiotherapy. Since these genes were identified using cases from the CGGA database, it is not surprising that these genes can better predict cases from the CGGA database than those from the TCGA database.
Nevertheless, some limitations of this study should be pointed out. First, the data were extracted from the CGGA database for the determination and validation of co-expressed modules rather than from the collection of original data. Moreover, there were no molecular and/or biochemical assays for further examination and validation of these hypothetical associations and/or interactions, which likely include false-positive results.
In summary, through the network construction of co-expressed genes by using WGCNA, we identified as well as validated a few critical hub genes that were regarded to be closely associated with the prognosis of glioma in Chinese patients, including KCNB1, UST, SOX8, KLHDC5, and HDAC4. Hence, these hub genes are potentially of great clinical significance and could possibly lead to promising approaches for personalized tumor therapy.
Table S1
| CGGA_ID | OS | Status | Radio | Chemo | IDH1.R32 | IDH2.R172 | TP53.1 | EGFR | ATRX | EZH2 |
|---|---|---|---|---|---|---|---|---|---|---|
| CGGA_1006 | 254 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| CGGA_1007 | 345 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| CGGA_1035 | 567 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| CGGA_1045 | 347 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_1072 | 84 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_1081 | 216 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_1145 | 239 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| CGGA_1177 | 449 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_1224 | 271 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| CGGA_1237 | 249 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_309 | 153 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_330 | 1,121 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_426 | 1,560 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_448 | 354 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| CGGA_483 | 271 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_488 | 435 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_564 | 679 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_578 | 376 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_604 | 381 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_616 | 952 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_643 | 331 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_658 | 372 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_669 | 607 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_676 | 376 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_678 | 386 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_727 | 1,023 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| CGGA_796 | 720 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| CGGA_802 | 681 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_808 | 309 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| CGGA_837 | 432 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| CGGA_842 | 234 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_D02 | 484 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_D03 | 423 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| CGGA_D11 | 827 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| CGGA_D37 | 965 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_D57 | 766 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_D62 | 514 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGA_1215 | 29 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| CGGA_300 | 2,199 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| CGGA_491 | 1,074 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| CGGA_545 | 555 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| CGGA_591 | 1,654 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| CGGA_632 | 572 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| CGGA_694 | 1,194 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| CGGA_747 | 970 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| CGGA_757 | 1,047 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGA_884 | 1,131 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGA_D47 | 1,048 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| CGGA_J023 | 1,028 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGA_1008 | 315 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CGGA_1119 | 436 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGA_247 | 609 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| CGGA_759 | 1,263 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
Acknowledgments
We thank Medjaden Bioscience for help with proofreading this article.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-492
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-492). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the review board of the local institution and the CGGA database of the Beijing Neurosurgical Institute at Capital Medical University. However, the requirement of written informed consent from the included patients with gliomas was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bo L, Wei B, Li C, et al. Identification of potential key genes associated with glioblastoma based on the gene expression profile. Oncol Lett 2017;14:2045-52. [Crossref] [PubMed]
- Sanai N, Berger MS. Recent surgical management of gliomas. Adv Exp Med Biol 2012;746:12-25. [Crossref] [PubMed]
- Senders JT, Harary M, Stopa BM, et al. Information-Based Medicine in Glioma Patients: A Clinical Perspective. Comput Math Methods Med 2018;2018:8572058. [Crossref] [PubMed]
- Kong Z, Yan C, Zhu R, et al. Imaging biomarkers guided anti-angiogenic therapy for malignant gliomas. Neuroimage Clin 2018;20:51-60. [Crossref] [PubMed]
- Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol 2017;134:505-12. [Crossref] [PubMed]
- Picca A, Berzero G, Di Stefano AL, et al. The clinical use of IDH1 and IDH2 mutations in gliomas. Expert Rev Mol Diagn 2018;18:1041-51. [Crossref] [PubMed]
- Qian M, Chen Z, Wang S, et al. PLEKHG5 is a novel prognostic biomarker in glioma patients. Int J Clin Oncol 2019;24:1350-8. [Crossref] [PubMed]
- Xu P, Yang J, Liu J, et al. Identification of glioblastoma gene prognosis modules based on weighted gene co-expression network analysis. BMC Med Genomics 2018;11:96. [Crossref] [PubMed]
- Wang HY, Wang W, Liu YW, et al. Role of KCNB1 in the prognosis of gliomas and autophagy modulation. Sci Rep 2017;7:14. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Chen J, Wang X, Hu B, et al. Candidate genes in gastric cancer identified by constructing a weighted gene co-expression network. PeerJ 2018;6:e4692. [Crossref] [PubMed]
- Chen L, Yuan L, Qian K, et al. Identification of Biomarkers Associated With Pathological Stage and Prognosis of Clear Cell Renal Cell Carcinoma by Co-expression Network Analysis. Front Physiol 2018;9:399. [Crossref] [PubMed]
- Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res 2010;1:274-8. [Crossref] [PubMed]
- Chen L, Yuan L, Wang Y, et al. Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma. Int J Biol Sci 2017;13:1361-72. [Crossref] [PubMed]
- Olar A, Wani KM, Alfaro-Munoz KD, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol 2015;129:585-96. [Crossref] [PubMed]
- Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol 2014;71:1319-25. [Crossref] [PubMed]
- Schlierf B, Friedrich RP, Roerig P, et al. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol 2007;33:621-30. [Crossref] [PubMed]
- Gomes MV, Borges KS, Moreno DA, et al. Abnormal methylation of histone deacetylase genes: implications on etiology and epigenetic therapy of astrocytomas. Anticancer Res 2011;31:1337-43. [PubMed]
- Cheng W, Li M, Cai J, et al. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J Neurooncol 2015;122:303-12. [Crossref] [PubMed]
- Marampon F, Megiorni F, Camero S, et al. HDAC4 and HDAC6 sustain DNA double strand break repair and stem-like phenotype by promoting radioresistance in glioblastoma cells. Cancer Lett 2017;397:1-11. [Crossref] [PubMed]
- Sferra R, Pompili S, Festuccia C, et al. The possible prognostic role of histone deacetylase and transforming growth factor beta/Smad signaling in high grade gliomas treated by radio-chemotherapy: a preliminary immunohistochemical study. Eur J Histochem 2017;61:2732. [Crossref] [PubMed]