
A feedback circuit of miR-34a/MDM4/p53 regulates apoptosis in chronic lymphocytic leukemia cells
Introduction
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in western countries with a mature B lymphocytes appearance (1) in morphology. The main mechanism was due to the deficiency in apoptosis signals such as TP53-induced glycolysis and apoptosis regulator (TIGAR) (2), checkpoint kinase 1 (Chk1) (3), particularly p53 associated pathways and so on. The conventional chemotherapy depends on normal functional p53 to induce the apoptosis of CLL cells.
It is noted that 50% of human cancers harbor defects, either deletion or mutation of p53 alleles (4), while over-expression of Murine double minute 2 (MDM2) and MDM4 were observed in cancers remaining wild type p53 (5-7). The fact of that the loss of MDM2 and MDM4 led to embryonic lethal mutation was first demonstrated in mice, while the mutation of MDMs could be rescued by the loss of p53, which indicated MDMs were essential and non-redundant in vivo (8-11).
MDM2 was proved to inhibit p53 by the similar RING domain coding protein in MDM2 and MDM4 under unstressed conditions (12). MDM2 is an E3 ubiquitin ligase, polyubiquitinating p53 and targeting p53 for degradation with its RING domain (13,14). MDM4 was first reported in 1996, which is a new member of MDM family and a structural homolog of MDM2 (15). MDM4 was reported to be over-expressed in appropriate 17.2% of human cancers (16). MDM4 is lack of ubiquitin ligase activity, and directly binding to p53 to inhibit transcriptional activity of p53. Besides, it could interact with MDM2 through the RING domain to stabilize MDM2 and to regulate the ubiquitination of p53 and MDM2 (17). Furthermore, Mancini et al. showed that MDM4 could act as a scaffold at mitochondria to bring p53 and BCL2 together and promote apoptosis (18). In previous study, MDM4 was over-expressed in CLL cells and might be a potential target in CLL therapy (19).
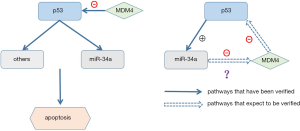
Induction of microRNA-34a (miR-34a) in concurrent with the decrease of MDM4 evoked by DNA damage has been observed in several cell lines (4). MiRNAs, a classical endogenously small non-coding RNAs, mediate gene silencing (20) by binding to the 3’UTR of mRNAs, which induces their destruction and/or inhibits translation. As bona fide p53 targets, miR-34 family, first reported in 2007 (21-23), is comprised of three processed miRNAs including miR-34a, miR-34b and miR-34c. MiR-34a has been reported to be 9-fold down-regulated in CLL cells with abnormal TP53 (24). The study conducted by Zenz et al. showed the low expression of miR-34a is correlated with a couple of causes including (I) deletion of 17p or mutation; (II) fludarabine-refractory disease especially in the absence of 17p deletion and (III) impaired response and apoptosis if DNA damage occurs (25). MiR-34a has been reported to be induced directly by p53 and promote apoptosis of CLL cells (21-23,26). Therefore, the aim of this study is to investigate potential modulating mechanisms of MDM4-P53-miR34a-interaction in primary CLL cells in vitro (Figure 1).
Methods
Clinical characteristics of enrolled patients
From February 2013 to October 2015, 33 enrolled patients were diagnosed with CLL at the First Affiliated Hospital of Nanjing Medical University. All cases, previously untreated or diagnosed with refractory and recurrent clinical course haven’t received any treatments for above 3 months (Table 1). Furthermore, we assessed prognosis associated factors including gender, age, Binet stage, IGHV mutational status, ZAP-70, CD38 and cytogenetics.
Table 1
| Characteristic | n (%) |
|---|---|
| Gender (n=33) | |
| Male | 22 (66.7) |
| Female | 11 (33.3) |
| Age (n=33) | |
| >60 | 13 (39.4) |
| ≤60 | 20 (60.6) |
| Binet stage (n=33) | |
| A | 11 (33.3) |
| B | 9 (27.3) |
| C | 13 (39.4) |
| IGHV (n=22) | |
| Unmutated (≤2% deviation from a germline) | 13 (39.4) |
| Mutated (>2% deviation from a germline) | 9 (27.3) |
| Not available | 11 (33.3) |
| ZAP-70 (n=27) | |
| >20% | 12 (36.3) |
| ≤20% | 15 (45.5) |
| Not available | 6 (18.2) |
| CD38 (n=26) | |
| >30% | 4 (15.4) |
| ≤30% | 22 (66.7) |
| Not available | 7 (21.2) |
| Cytogenetics (n=33) | |
| Deletion in 17p13 (n=25) | 7 (21.2) |
| P53 mutation (n=13) | 3 (9.1) |
Ethic statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committees of the First Affiliated Hospital of Nanjing Medical University (No. 2011-SRFA-031) and informed consent was taken from all the patients. All of the analyses were performed in accordance with the principle of relevant guidelines and regulations.
Cell culture
Peripheral blood samples were obtained from 33 patients with CLL. Mononuclear leucocytes were isolated from peripheral blood by Ficoll-isopaque centrifugation and above 90% of cells were CD19 positive according to flow cytometry. The cells were re-suspended in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum in a 5% CO2-humidified atmosphere at 37 °C for 24 h.
Luciferase reporter assays
According to the bioinformatic approaches, there are 4 miRNA-34a binding domains in the 3’UTR (Figure 2), even if Mandke et al., 2012 pointed out that MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame (27). We take advantage of a commercial cell line, 293T cells (HEK-293T, Human embryonic kidney cell line) (ATCC Cat# CRL-3216, RRID: CVCL_0063) to verify our hypothesis in luciferase reporter assays based on the limitation of CLL cell lines in vitro. The 3’UTR and exon 11 of MDM4 containing the putative binding site of miR-34a were established and cloned in the firefly luciferase-expressing vector psiCHECK-2 (psiCHECK-2 Vector is designed to provide a quantitative and rapid approach for optimization of RNA interference) (Promega Corporation) respectively, and then named psiCHECK-2-MDM4-UTR or the psiCHECK-2-MDM4-exon. Cells were seeded in 48-well plates for 24 h before transfection and were transiently transfected with either the psiCHECK-2-MDM4-UTR or the psiCHECK-2-MDM4-exon together with miR-34a mimics or miR-Ctrl combined with lipofectamine 2000 (Invitrogen, USA). After 48 h, cells were harvested and detected for luciferase activities by the Dual-luciferase Reporter Gene assay kit (Promega, Beijing, China). We conducted three culture repeats for the average and ensured accuracy. Details and procedure were provided as supplemental experimental procedures (two PDF file A6069-1 and A6069-2 & Luciferase reporter assays, available at: http://fp.amegroups.cn/cms/3e457ba8f49a582ca4e35f11426a7c39/tcr-20-1710-1.docx; http://fp.amegroups.cn/cms/5f082ba7df2c4fd04904502ede5825b2/tcr-20-1710-2.docx; http://fp.amegroups.cn/cms/2b5b268be17cd131ae1f5cc40bc1e2f0/tcr-20-1710-3.docx).
Luminescence in situ hybridization (FISH)
FISH and PCR analysis were performed on the samples 25 from 33 CLL patients. The following fluorescent labeled probes were used in interphase FISH: LSI MYB (6q23), LSI ATM (11q22), LSD13S319 (13q14), LSI IGHC/IGHV (14q32), LSI p53 (17p13) and CEP12 (centromere 12) (all probes were purchased from Vysis, Downers Grove, IL, USA).
Flow cytometry
Flow cytometric analysis of CD38 and ZAP-70 was performed on fresh peripheral blood or bone marrow samples stained with CD5-FITC, CD19-ECD, CD38-PE (clone HB-7; BD Biosciences) and ZAP-70-PE (clone 1F7.2; Caltag). Isotype controls were run with each sample to distinguish positive cells from negative cells. The cut off point for CD38-positive and ZAP-70-positive in CLL cells was 30% and 20%, respectively.
Transfection
The isolated cells were predominantly clonal B cells (>90% CD5+CD19+), as assessed by flow cytometry (FACScan, Becton Dickinson, San Jose, CA, USA). Freshly isolated CLL cells were seeded in six-well plates (5×106 cells/well). After 6 h, they were treated with mixture of 3 µL lipo2000 (Invitrogen) and 30 pmol miR-34a mimics (5'-UGGCAGUGUCUUAGCUGGUUGU-3', 5'-AACCAGCUAAGACACUGCCAUU-3')or negative control (NC) (5'-UUCUCCGAACGUGUCACGUTT-3', 5'-ACGUGACACGUUCGGAGAATT-3'), 0.8 µg short hair MDM4 (shMDM4) (5'-GCTGCTGATACTGAACAAACA-3', 5'-GCAACTCAGTGGAATTCTTGG-3') or sh-negative control (shNC) (5'-GTTCTCC GAACGTGTCACGT-3') (Genephama, Shanghai, China) and then cultured in RPMI-1640 medium supplemented with 10% FCS in a humidified atmosphere containing 5% CO2 at 37 °C. The CLL cells were harvested for RNA and protein extraction after 24 h.
Detection of apoptosis
The apoptosis rate was detected 24 hours after the transfection. Use Annexin V/PI double staining to detect apoptosis. Alive cells were Annexin V-positive (–) and PI-negative (–), necrotic cells were Annexin V-positive (+) and PI-negative (+), while apoptotic cells were Annexin V-positive (+) and PI-negative (–). In our research, the percentage of Annexin V-positive (+) and PI-negative (–) cells in all including alive cells, necrotic cells and apoptotic cells was defined as apoptosis rate using the Annexin V kit (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time RT-PCR
miRNA qRT-PCR was performed according to the manufacturers’ recommendations. Total RNA was isolated from CLL cells by Trizol (TaKaRa, Japan). After isolation of RNA, 1 µg of total RNA from each sample was reverse-transcribed using the Reverse Transcription Reagents (Applied Biosystems). Reverse transcriptase reactions contained: 1 µg of purified total RNA, 5×M-MLV buffer (Invitrogen, USA) 4.0 µL, M-MLV (200 U/µL, Invitrogen, USA) 1.0 µL, DTT 1.0 µL, stem-loop RT primer (10 µmol/L) 1.0 µL, RNase Inhibitor (40 U/µL) 0.5 µL, dNTP (10 mmol/L) 1.0 µL, 20 µL reactions were performed with the following thermal cycling parameters: 60 min at 42 °C, 5 min at 85 °C and then held at 4 °C. Reactions for qRT-PCR were conducted in duplicate, using the Applied Biosystems ABI 7300 Real Time PCR system. Each reaction mixture was contained of 3 µL of RT product and SYBR GREEN master (Roche, Switzerland) 10 µL, 0.5 µM of each primer (Table 2), and deionized water up to a total volume of 20 µL. Reactions were run with the following thermal cycling parameters: 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s (denaturation) and 60 °C for 30 s, melting curve stage of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. Each sample was normalized based on its endogenous U6 RNA content.
Table 2
| Primer | Sequence (5'-3') |
|---|---|
| U6 | RT AACGCTTCACGAATTTGCGT |
| Forward CTCGCTTCGGCAGCACA | |
| Reverse AACGCTTCACGAATTTGCGT | |
| miR-34a | RT CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACCAG |
| Forward ACACTCCAGCTGGG TGGCAGTGTCTTAGCT | |
| Reverse TGGTGTCGTGGAGTCG | |
| MDM4 | Forward TGGGAGAACTACTGGGACGT |
| Reverse TCCTGTGCGAGAGCGAGAGT | |
| p53 | Forward ATGAGCGCTGCTCAGATAGC |
| Reverse TCAAAGCTGTTCCGTCCCAG | |
| β-actin | Forward AGCGAGCATCCCCCAAAGTT |
| Reverse GGGCACGAAGGCTCATCATT |
The levels of putative target mRNA (MDM4, p53) were also evaluated by real-time PCR. For the PCR analysis, β-actin was used as an internal control. The PCR primers for β-actin, MDM4 and p53 were designed using the Primer 5.0 (Table 2). In a total volume of 20 µL of reaction mixture, 6 µL of complementary DNA templates were mixed with 10 µL of SYBR GREEN master (Roche, Switzerland) according to the manufacturer’s instruction and each pair of primers at a final concentration of 100nM. Cycle conditions for MDM4, p53 and β-actin were one cycle for 10 min at 95 °C, 35 cycles for 30 s at 95 °C, 30 s at 58 °C, 30 s at 72 °C, and finally, 1 cycle for 15 s at 95 °C, 1 min at 57 °C and 15 s at 95.3 °C. The threshold cycle (Ct) was defined as the fractional cycle number at which the luminescence passes the fixed threshold, and each sample was normalized based on its endogenous β-actin RNA content and was assayed in duplicate.
Setting 3 repetitions for each hole, avoiding light on ice. Each specimen simultaneously amplified the target gene and U6 internal reference gene. The experiment uses the following formula to calculate the relative expression of the target gene: the relative expression of the target gene miRNA = 2 (U6-miRNA) (U6: Ct value of the internal reference gene U6). Relative expression of target gene MDM4 =2 (β-actin-MDM4) (β-actin: Ct value of internal reference gene β-actin). Relative expression of target gene p53= 2 (β-actin-p53) (β-actin: Ct value of internal reference gene β-actin).
IGHV mutational status was determined by PCR-based DNA sequencing from DNA extracted from peripheral blood or bone marrow samples, with a cut off value of that more than 2% germline divergence was defined as “mutated”.
Statistical analysis
All statistical analyses were performed using the SPSS 20.0 and GraphPad Prism 5.0. Differences between groups were analyzed using Mann-Whitney U test P<0.05 was considered statistically significant. All statistical testing was performed taking into account that paired statistical tests should be used in all tests. An effect was considered statistically significant at P<0.05. ΔCt was calculated by subtracting the Ct of reference genes (U6 small nuclear RNA and β-actin) from the Ct of the genes of interest. The relative amount of miRNAs and MDM4, p53 mRNAs was calculated by the equation 2-ΔCT. MiRNAs expression levels were compared by Mann-Whitney independent t-test.
Results
Characteristics of patients
The clinical characteristics of 33 CLL patients are summarized in Table 1. A total of 33 patients were included with 22 male and 11 female patients. The median age was 56 (range, 23–75) years old. According to the Binet clinical staging system, 11 patients (33%) were classified as Binet stage A, 9 (27%) as Binet stage B, and 13 (39%) as Binet stage C. There were 7 patients containing a deletion of 17p13 and 3 patients harboring a TP53 mutation. Only part of the cohort was investigated for 17p deletion (25/33, 7 cases were positive) and TP53 mutation (13/33, 3 cases were positive). Not all patients were first treated or diagnosed in our hospital and some of them refused to accept the detection of 17p deletion and TP53 mutation.
Luciferase reporter assay
The luminescence of psiCHECK-2-MDM4 EXON 11 can be effectively inhibited by has-miR-34a (inhibition rate: 24%, P=0.007, Figure 3) (Supplemental File: Luciferase reporter assays) while the luminescence of other binding region in 3’UTR could not be obviously decreased (P=0.829), which indicate that miR-34a can binding to MDM4 through domain of exon 11 instead of 3’UTR in 293T cells even if Mandke et al., 2012 pointed out that MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame (27).
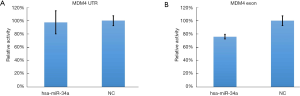
The luminescence of psiCHECK-2-MDM4 EXON 11 can be effectively inhibited by has-miR-34a (inhibition rate: 24%, P=0.007) while that of psiCHECK-2-MDM4 UTR can not be inhibited by has-miR-34a (inhibition rate: 3%, P=0.829). Lines represented the mean ± SD.
Changes of mRNAs expression after transfection
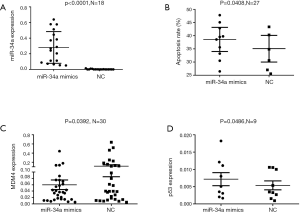
Hsa-miR-34a mimics and miR-NC were transfected into primary CLL cells in vitro respectively and harvested after 24 h for evaluating expression of miRNA by RT-PCR. And the changes of mRNAs expression were presented in both Figures 4 and 5. The results showed that median miR-34a mRNA level in the group transfected with hsa-miR-34a and miR-NC was 0.254 (95% CI, 0.180–0.382) and 0.0004 (95% CI, 0.0002–0.004) respectively (P<0.005) (Figure 4A, line presented mean), which indicates the transfection of Hsa-miR-34a mimics was efficient. Owing to the limitation of samples, 18 cases of 32 samples were analyzed for apoptosis of hsa-miR-34a and miR-NC.

ShMDM4 and sh-NC were transfected into primary CLL cells in vitro respectively and harvested after 24 h for evaluate expression of MDM4 RNA by RT-PCR. The results showed that median MDM4 mRNA level in the group transfected with shMDM4 and sh-NC was 0.033 (95% CI, 0.031–0.063) and 0.06291 (95% CI, 0.048–0.202) respectively (P=0.031) (Figure 5A, line presented mean), which indicates the transfection of ShMDM4 was efficient.
Apoptosis rate
Our pre-experiment found that the number of chronic lymphocytes that died after 48 h was high in several cases. Considering the fragile CLL cells in vitro, we made the choice of 24 h instead of 48 h to evaluate the apoptotic process. The proportion of apoptosis cells was increased after CLL cells were transfected with hsa-miR-34a mimics or shMDM4 compared with miR-NC (Figure 4B, plot represented Mean with SEM) and shNC (Figure 5B, line at Mean with SEM) respectively. The median apoptosis rate of CLL cells after transfected with hsa-miR-34a mimics was 35.6% (95% CI, 29.2–48.0%) while the median apoptosis rate of miR-NC was 25.6% (95% CI, 24.7–45.5%) (P=0.041) (Figure 4B, plot represented Mean with SEM), the median apoptosis rate of CLL cells transfected with shMDM4 was 27.4% (95% CI, 25.8–47.0%) while that of shMDM4-NC was 25.8% (95% CI, 21.1–43.5%) (P=0.015) (Figure 5B, line at Mean with SEM). According to our research, increased expression of miR-34a or shMDM4 can promote apoptosis of CLL cells.
Correlation between miRNA and predicted target genes
We analyzed correlation among expression of MDM4, TP53 and miR-34a mRNA. As the effect of miR-34a transfection on MDM4 expression is showed in Figure 4C, line at Mean with SEM, Primary CLL cells transfected with hsa-miR-34 mimics showed a lower MDM4 mRNA expression compared with NC (P=0.039). The median mRNA expression of MDM4 was 0.037 (95% CI, 0.029–0.091) after transfected with miR-34a mimics while the median mRNA expression in miR-NC was 0.040 (95% CI, 0.055–0.174) (P=0.039) (Figure 4C, line at mean with SEM). MDM4 could be inhibited by upregulating miR-34a depending on our results. A negative correlation between down-regulated expression of MDM4 mRNA and TP53 mRNA was observed (P=0.029) (Figure 5C, line presented mean). The median expression of TP53 was 0.005 (95% CI, 0.003–0.008) after being transfected with shMDM4 while the median expression of TP53 in shNC group was 0.004286 (95% CI, 0.003–0.007). TP53 could be indirectly upregulated by downregulating MDM4 according to our research. Interestingly, expression of TP53 in mRNA after transfected with miR-34a mimics was 0.005 (95% CI, 0.003–0.012) while the mRNA expression of TP53 in NC group was 0.003 (95% CI, 0.002–0.008). TP53 expression was observed increased after upregulating miR-34a compared to NC group (P=0.049) (Figure 4D, line presented mean), which might due to indirectly downregulating MDM4 or much more other signal pathways. These results suggest that MDM4 could be suppressed by miR-34a in CLL cells while an indirect negative correlation between MDM4 and TP53 mRNA expression was detected, which is consistent with pathways showed in Figure 1.
Discussion
CLL is conventionally treated by routine chemotherapy depending on its normal functional TP53. Increased expression of MDM4 associated with aberrant TP53 genotypes indicated poor prognosis. Over-expression of MDM4 was constantly found in several malignancies including CLL and Mantle cell lymphoma (19,28). Interestingly, expression of MDM4 was increased in part of CLL patients with deletion or mutation of TP53 and its expression was associated with staging and deletion of ATM (29). TP53 plays a crucial role in inhibiting tumorigenesis by stabilizing the homeostasis of genome (30). Some targeting genes can be inhibited by miR-34a in post-transcription period to modulate the formation and development of tumors (31). Besides, miR-34a was proved to show a negative correlation with MDM4 in the specific tissue from human lung adenocarcinoma (32). MiRNAs encoding by endogenous genes take actions in post-transcriptional level through targeting at specific mRNAs (20) linked with the process of CLL (30-35). It is suggested that miR-34a may play a crucial role in the pathogenesis of CLL. Considering of the difficulties of primary CLL cells culture and transfection in vitro and limitation of volume in peripheral blood samples, we adopt one kind of method like annexin-V and PI staining to detect apoptosis of our samples in time without a second method like detection of cleaved PARP or caspases to validate these results to enhance credibility of our research. On the basis of the results that upregulated expression of miR-34a resulted in increased apoptosis of CLL cells, decreased expression of MDM4 accompanied with increased expression of TP53, it was proved that miR-34a could indirectly modulate expression of MDM4, and then modulate TP53. We firstly guarantee the enough samples for the experiment of detection of apoptosis rate, expression of mRNA and miRNA by RT-PCR, and didn’t have enough samples for further detection of protein level. Treatment with miR-34a mimics or sh-MDM4 for 48–72 h ex-vivo might give better results with respect to apoptosis if CLL cells overcame their fragility ex-vivo.
Based on our hypothesis, we hope that the increased apoptosis rate induced by miR-34a or MDM4 modulation in primary CLL is TP53-mediated. According to our research, a total of 32 samples transfected with miR-34a mimics and NC were used for analysis efficiency of transfection of miR-34a, and 27 cases were detected for apoptosis rate. And all these samples with TP53 aberrations (TP53 mutation: 1 case, TP53 mutation + 17p deletion: 2 case, 17p deletion: 3 case) didn’t showed obvious increased apoptosis rate after transfection of miR-34a. We tested the kind of mutation of the three patient with mutTP53 and found the ability of inactivating partially while we thought 17p deletion played a much more important role. Unfortunately, we have not detected the percentage of cells carrying del17p/TP53 mut to clarify if they were present at clonal or subclonal level. Besides, a total of 28 samples were transfected with MDM4 and sh-NC for analysis efficiency of transfection of MDM4, and 24 cases were detected for apoptosis rate. And 4 cases harboring p53 aberrations (17p deletion: 1 case, 17P deletion + p53 mutation: 2 case, p53 mutation: 1 case) transfected with shMDM4 didn’t showed increased apoptosis rate after transfection of shMDM4 while 2 cases harboring 17p deletion increased apoptosis rate. On the basis of above results, we think miR-34a and MDM4 could modulate apoptosis depend on functional TP53 signaling pathway. Besides, upregulated MDM4 may partly regain TP53 function especially in p53 aberrations. Considering the deficient number of cases with TP53 aberrations, it needs much more exploration.
The luminescence of psiCHECK-2-MDM4 EXON 11 can be effectively inhibited by has-miR-34a. MiR-34a could directly modulate the expression of MDM4 by binding to the region of MDM4 exon 11, which provides direct evidence for the exploration of miR-34a/MDM4 associated pathway and a new idea for the treatment of CLL. Besides, miR-34a couldn’t bind to 3'UTR of MDM4 while it could bind to the protein coding region (opening reading frame) above 3'UTR. Even if so many miRNAs take effects on target genes through its 3'UTR, a little miRNA could bind to some coding regions. DNMT1 could be regulated by miR-148 through coding region (36) and HNF4A could be modulated by miR-34a through coding region (37).
Furthermore, decreased expression of MDM4 could cause gained apoptosis of CLL cells and increased expression of TP53 mRNA. In our manuscript, 5 cases of harboring TP53 aberrations (17p deletion: 4 cases, 17P deletion + p53 mutation: 1 case) were transfected with miR-34a mimics and 7 cases harboring TP53 aberrations (17p53 deletion: 4 cases, 17P deletion + TP53 mutation: 2 case, TP53 mutation: 1 case) were transfected with shMDM4. MDM4 and p53 at protein level by western blotting before and after transfection may strengthen our conclusions. The relevant limitations of this study that MDM4 silencing results in a similar phenotype does not demonstrate, but simply suggests that the effects of miR-34a might be mediated by MDM4 reduction.
Jones et al. pointed out the combination of an human homolog of the murine double minute protein-2 (HDM-2) with MI-63 and ribonucleotide reductase inhibitor like gemcitabine shed a light on patients with relapsed wt TP53 MCL (38). Nutlin-3, as an inhibitor of MDM family members, plays an important role in inducing apoptosis of MCL and HL by efficiently activating TP53. But we haven’t found the similar phenomena in other lymphomas such as FL, DLBCL, MZL, specially HBV+ or EBV+ lymphoma. It provides us a new direction to explore the potential feedback loop in other lymphomas.
Conclusions
Although MDM4 and miR-34a are vital partials in TP53 pathways, the reciprocally modulating mechanisms among them haven’t been clarified clearly in hematological malignancies (39). It is constantly proved that MDM family members can modulate p53 in transcriptional and posttranscriptional levels via binding TP53, sealing the active transcriptional domain of TP53 and realizing inhibiting transcription and function of mitochondria. MiR-34a prevents such genes including MDM4 in post-transcriptional level to enhance the activity of TP53 and decrease the degradation of TP53. MDM4 was proved to be directly inhibited by miR-34a according to luciferase reporter assay which is different from Mandke et al.’s research (27) and be indirectly upregulated through downregulating miR-34a based on results of transfection. TP53 was proved to be indirectly upregulated by inhibiting MDM4 or upregulating miR-34a. Apoptosis rates of CLL cells were detected to be increased after transfection with miR-34a mimics or shMDM4. Above all, we draw a conclusion that regaining the balance among p53/miR-34a/MDM4 pathway may shed a light on the treatments of hematological malignancies worthy to be further explored and expounded.
Acknowledgments
We thank all members of our group for their efforts and enthusiasm.
Funding: The study got its vigorously supported funds from
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1710
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1710). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committees of the First Affiliated Hospital of Nanjing Medical University (No. 2011-SRFA-031) and informed consent was taken from all the patients. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zenz T, Mertens D, Küppers R, et al. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010;10:37-50. [Crossref] [PubMed]
- Hong M, Xia Y, Zhu Y, et al. TP53-induced glycolysis and apoptosis regulator protects from spontaneous apoptosis and predicts poor prognosis in chronic lymphocytic leukemia. Leuk Res 2016;50:72-7. [Crossref] [PubMed]
- Zemanova J, Hylse O, Collakova J, et al. Chk1 inhibition significantly potentiates activity of nucleoside analogs in TP53-mutated B-lymphoid cells. Oncotarget 2016;7:62091-106. [Crossref] [PubMed]
- Markey M, Berberich SJ. Full-length hdmX transcripts decrease following genotoxic stress. Oncogene 2008;27:6657-66. [Crossref] [PubMed]
- Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 2010;24:1580-9. [Crossref] [PubMed]
- Momand J, Jung D, Wilczynski S, et al. The MDM2 gene amplification database. Nucleic Acids Res 1998;26:3453-9. [Crossref] [PubMed]
- Oliner JD, Kinzler KW, Meltzer PS, et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992;358:80-3. [Crossref] [PubMed]
- Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206-8. [Crossref] [PubMed]
- Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527-38. [Crossref] [PubMed]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [Crossref] [PubMed]
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92-5. [Crossref] [PubMed]
- Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 2010;20:299-309. [Crossref] [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [Crossref] [PubMed]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997;420:25-7. [Crossref] [PubMed]
- Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996;15:5349-57. [Crossref] [PubMed]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006;6:909-23. [Crossref] [PubMed]
- Chua JS, Liew HP, Guo L, et al. Tumor-specific signaling to p53 is mimicked by Mdm2 inactivation in zebrafish: insights from mdm2 and mdm4 mutant zebrafish. Oncogene 2015;34:5933-41. [Crossref] [PubMed]
- Mancini F, Moretti F. Mitochondrial MDM4 (MDMX): an unpredicted role in the p53-mediated intrinsic apoptotic pathway. Cell Cycle 2009;8:3854-9. [Crossref] [PubMed]
- Bo MD, Secchiero P, Degan M, et al. MDM4 (MDMX) is overexpressed in chronic lymphocytic leukaemia (CLL) and marks a subset of p53wild-type CLL with a poor cytotoxic response to Nutlin-3. Br J Haematol 2010;150:237-9. [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007;17:1298-307. [Crossref] [PubMed]
- Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007;26:745-52. [Crossref] [PubMed]
- He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130-4. [Crossref] [PubMed]
- Mraz M, Malinova K, Kotaskova J, et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 2009;23:1159-63. [Crossref] [PubMed]
- Zenz T, Mohr J, Eldering E, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 2009;113:3801-8. [Crossref] [PubMed]
- Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007;26:731-43. [Crossref] [PubMed]
- Mandke P, Wyatt N, Fraser J, et al. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One 2012;7:e42034. [Crossref] [PubMed]
- Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005;353:1793-801. [Crossref] [PubMed]
- Liu L, Fang C, Dong HJ, et al. mRNA levels detected by real-time quantitative RT-PCR in chronic lymphocytic leukemia and their clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2011;19:1145-9. [PubMed]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2010; [Crossref] [PubMed]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010;17:193-9. [Crossref] [PubMed]
- Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 2014;28:438-50. [Crossref] [PubMed]
- Rossi S, Shimizu M, Barbarotto E, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 2010;116:945-52. [Crossref] [PubMed]
- Asslaber D, Piñón JD, Seyfried I, et al. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood 2010;115:4191-7. [Crossref] [PubMed]
- Zhu DX, Zhu W, Fang C, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 2012;33:1294-301. [Crossref] [PubMed]
- Duursma AM, Kedde M, Schrier M, et al. miR-148 targets human DNMT3b protein coding region. RNA 2008;14:872-7. [Crossref] [PubMed]
- Takagi S, Nakajima M, Kida K, et al. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 2010;285:4415-22. [Crossref] [PubMed]
- Jones RJ, Baladandayuthapani V, Neelapu S, et al. HDM-2 Inhibition Suppresses Expression of Ribonucleotide Reductase Subunit M2, and Synergistically Enhances Gemcitabine-Induced Cytotoxicity in Mantle Cell Lymphoma. Blood 2011;118:4140-9. [Crossref] [PubMed]
- Navarro F, Lieberman J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS One 2015;10:e0132767. [Crossref] [PubMed]



