Emodin inhibits the proliferation and migration of B16F10 cells and induces their apoptosis
Introduction
Cutaneous melanoma (CM) is a skin tumour derived from abnormal melanocytes that is prone to migrate in the early stage. It accounts for approximately 75% of skin cancer-related deaths (1). Recently, the incidence of CM is one of the most rapidly increasing among cancers worldwide. The increased incidence varies among populations, ranging from 3% to 7% (2). Studies from Europe and the United States reports increases in the incidence of CM (3,4).
The mortality of melanoma has also increased in men older than 65 years. This finding may be attributed to their unique biology and/or an altered host immune response (5). Previously, treatment options were limited by modest response rates in patients with advanced-stage melanoma. Despite targeted therapy and immunotherapy raising the response rate in recent years, the long-term survival rate remains low. Therefore, the discovery of efficient therapeutics will be key for treating CM to improve overall survival.
Emodin, extracted from the rhizomes of various Chinese herbal medicines, such as rhubarb, rheum and polygonum, is a natural anthraquinone derivative with the formula C15H10O5. The chemical name of emodin is 1,3,8-trihydroxyanthraquinone. It forms an orange-yellow long needle-like crystal. In recent years, many studies have confirmed that emodin has many biological effects, such as anti-inflammatory, antibacterial, and immune regulation effects (6-8). With further research on its antitumour activity, emodin was found to inhibit lung cancer, breast cancer and other tumours by inhibiting tumour cell proliferation and promoting apoptosis (9,10).
This study mainly investigated the effect of emodin on the proliferation, migration, and apoptosis of the B16F10 mouse melanoma cell line. The goal was to further understand the mechanism of apoptosis and provide experimental evidence for the treatment of melanoma with emodin.
Methods
Materials
Emodin and dimethyl sulfoxide (DMSO) were purchased from Sigma. DMEM, foetal bovine serum, a CCK-8 kit, and 0.25% EDTA-trypsin were purchased from Gibco (Thermo Fisher Scientific, Inc). A TUNEL kit and DAPI staining kit were purchased from Nanjing Kaiji Biotechnology Co., Ltd.; a penicillin and streptomycin solution were purchased from United Bioresearch, Inc. Six-well cell plates and 96-well cell plates were purchased from Shanghai Jiayuan Biotech Co., Ltd. The polyvinylidene fluoride (PVDF) membranes were purchased from Beijing Solarbio Science & Technology Co., Ltd. The antibodies against β-actin (rabbit anti-mouse RRID: AB_2242334) and caspase-3 (RRID: AB_331439) were purchased from Cell Signalling Technology, Inc. The secondary fluorescence antibody (polyclonal goat anti-rabbit RRID: AB_2801422) was purchased from Yeasen Biotech Co., Ltd.
Cell line and cell culture
Mouse melanoma B16F10 cells (RRID: CVCL_0159) were purchased from the Cell Bank of the Chinese Academy of Sciences. B16F10 cells were grown in DMEM (pH=7.4) with 10% foetal bovine serum and penicillin (final concentration, 100 U/L) and streptomycin (final concentration, 100 mg/L). The cells were cultured in an incubator with 95% air and 5% CO2 with saturated humidity at 37 °C. The cells in the logarithmic growth phase were selected for experiments and digested with EDTA-trypsin when their confluence reached 80–90%. The cells were seeded into six 96-well plates. The medium was changed every 2 days.
Preparation of emodin
Emodin was dissolved in DMSO at a concentration of 0.2 mmol/L as a stock solution and stored in a refrigerator at −20 °C. The stock solution was diluted to a desired concentration in DMEM medium at the time of use. The mass fraction of DMSO in the emodin solution was less than 0.1%.
Cell proliferation assay
The cells in the logarithmic growth phase were washed twice with PBS, and 0.25% trypsin was used to detach the cells at room temperature, and the cells was transferred to a centrifuge tube. After resuspending the cells, the cells were counted by a haemocytometer under the microscope and seeded into a 96-well plate at 104 cells/well. The seeded cells were cultured for 24 h. The 0.1% DMSO group was simultaneously prepared as a control, and the experimental group was treated with 20, 40, 60, or 80 µM emodin for 24 h. The blank zero group was treated with equal amounts of PBS. After adding 10 µL of CCK-8 reagent to each well, the cells were incubated for 4 h in an incubator. The absorbance was measured at a wavelength of 450 nm, and the viability of the B16F10 cells in different concentrations of emodin was calculated.
Cell scratch test
The cells in the logarithmic growth phase were added to 6-well plates. Three parallel lines were drawn as a marker underneath the 6-well plate. When the confluence reached 100%, a 100 µL pipette tip was used to place a scratch vertically and slowly into the 6-well plate. The wells were washed with PBS three times, and serum-free medium with 20, 40, 60, and 80 µM emodin was added. Serum-free (0.1% DMSO) medium was simultaneously added to a well as the control. The cells were observed under an inverted microscope at 0 h and 24 h, and ImageJ (RRID:SCR_003070) was used to calculate the migration rate.
Transwell test
Cells in the logarithmic growth phase were added to the upper chamber at a density of 2×105/mL (150 µL) and treated with 20, 40, 60, or 80 µM emodin in serum-free medium for 24 h. The control group was treated with 0.1% DMSO. Then, 800 µL of medium containing 10% foetal bovine serum was added to a 24-well plate placed in the lower chamber. The 24-well plate was placed in an incubator for 24 h. The upper chamber liquid was blotted and carefully wiped with a cotton swab after 24 h. The cells on the surface of the upper chamber were removed, and the bottom surface was fixed with 4% paraformaldehyde and stained with crystal violet for 20 min. The number of migrating cells was counted in different fields of view under a microscope. Five visual fields were randomly selected for statistical analysis, and the differences between the groups were statistically analysed.
TUNEL assay
Cell slides were washed with deionized water and sterilized for use. B16F10 cells were seeded onto 6-well plates at a density of 2×105/well and cultured overnight. After the cells were attached, emodin (60 µM) was added, and incubation was continued for 24 h. The cells attached to the cell slides were naturally dried and fixed with 4% paraformaldehyde for 30 min and washed with PBS 3 times. The TUNEL reaction solution was prepared according to the instructions, and the whole process was performed on ice. The reaction solution was added dropwise to each cell slide to allow the reaction solution to completely cover the cell area. The reaction was incubated at 37 °C for 1 h in the dark and washed 3 times with PBS for 5 min each time. The DAPI solution was diluted to 1 mg/L and then added dropwise to the cell slide. The cells were then incubated at 37 °C for 15 min in the dark and then washed 3 times with PBS for 5 min each time. The image was observed under an inverted fluorescence microscope after a quenching agent was added, and the surrounding area was devoid of cells. Three fields of view were randomly selected and photographed under a laser confocal microscope.
Western blot analysis
Proteins were collected and separated through SDS-polyacrylamide gel electrophoresis from cells that had reached 80–90% confluence and treated with 20, 40 and 60 µM emodin for 24 h. The control group was treated identically. The separated proteins were transferred to a PVDF membrane with the application of 90 V and 200 mA for 60 min. Next, PVDF membranes were incubated overnight in a diluted antibody solution (rabbit anti-mouse caspase-3 and anti-β-actin; 1:3,000) at 4 °C. The blots were then incubated with the secondary antibody (1:3,000) for 1 h at 37 °C. The results were detected using an Odyssey infrared imaging system.
Statistics
After all the experiments were repeated 3 times, the measurement data were expressed as the means ± standard deviation (), and the data were processed by SPSS 19.0 statistical software (RRID:SCR_002865). The two groups were compared by t-test, and P<0.05 was used to indicate a statistically significant difference.
Results
Effect of emodin on the proliferation of the B16F10 cells
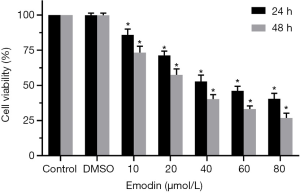
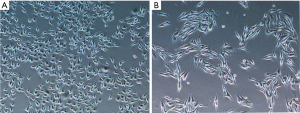
B16F10 cells were treated with different concentrations of emodin (20, 40, 60, 80 µM) for 24 h and 48 h, and the viability of the B16F10 cells showed a decreasing trend. There was a significant difference compared with the control group (P<0.05) (Figure 1). As the emodin concentration increased, the cell viability rate decreased, showing that emodin inhibited the proliferation of B16F10 cells in a concentration-dependent manner. Changes in the cell morphology of the B16F10 cells treated with 60 µM emodin for 24 h were observed under an inverted microscope (Figure 2). After treatment with 60 µM emodin, the number of B16F10 cells decreased, and the morphology of the cells began to show reticular dendritic structures. Some cell volumes decreased, and the shape became round.
Effect of emodin on the migration of the B16F10 cells
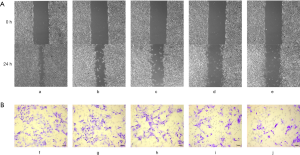
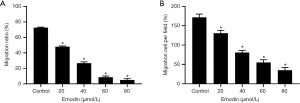
After treatment of the B16F10 cells with gradient concentrations of emodin (20, 40, 60, and 80 µM) for 24 h, cell migration changes were observed under an inverted microscope (Figure 3A). B16F10 cells were treated with the same gradient concentration of emodin solution for 24 h in the Transwell test. The number of cells migrating to the bottom surface of the membrane decreased in a concentration-dependent manner (Figure 3B). The migration ratio of the cells the experimental group was significantly reduced compared with that of the cells in the control group (P<0.05) (Figure 4A). The number of cells migrating to the bottom surface of the membrane was significantly different from that of the control group (P<0.05) (Figure 4B).
Effect of emodin on the apoptosis of the B16F10 cells
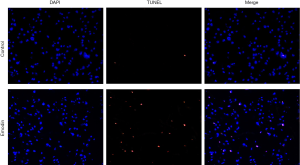
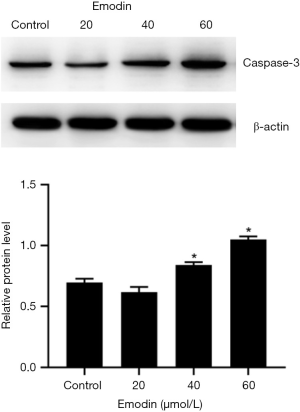
Compared with those of the control group, the B16F10 cells treated with emodin at a concentration of 60 µM for 24 h showed an increased number of fluorescently stained cells (Figure 5). Caspase-3 has an important impact on apoptosis. In our study, caspase-3 was detected and analysed by western blotting, which indicated that the expression levels of caspase-3 had increased in cells treated with 40 µM and 60 µM emodin (Figure 6). These results confirmed that emodin may promote B16F10 cell apoptosis.
Discussion
The incidence of CM has been one of the most rapidly increasing among cancers worldwide. The proliferation and invasive ability of CM are the main factors leading to early lymphatic and distant metastasis. The poor chemotherapy effect on melanoma has led to a 5-year survival rate of patients with advanced melanoma that is still low (11). It is gratifying that with further research of melanoma performed in recent years, some advancements in targeted therapy and immunotherapy have been achieved (12,13). Recently, several studies have shown that emodin can inhibit tumour growth by inhibiting tumour cell proliferation and inducing apoptosis (14-16). Emodin can also synergize with various antitumour drugs to reduce tumour resistance (17). Aloe-emodin, reported as a potential anticancer agent, has a molecular structure similar to that of emodin. Aloe-emodin can reduce tumour cell viability in a dose-dependent manner and inhibit the metastasis of tumours formed by B16F10 cells (18). The antitumour effects of emodin may similar to drugs with a similar anthraquinone structure (Figure 7). Additionally, emodin can upregulate the expression of caspase-3 to induce the apoptosis of HeLa cells via the mitochondrial pathway and upregulate the expression of Fas and FasL to induce apoptosis through the extracellular pathway (19). Therefore, we explored the effect of emodin on B16F10 cells to provide evidence of its potential clinical application in the treatment of melanoma.
In this study, it was found that the viability of the B16 cells decreased significantly with increasing emodin concentration, indicating that emodin inhibited the proliferation of B16F10 cells in a concentration-dependent manner. The changes in the morphology of B16F10 cells treated with 60 µM emodin suggest the possible induction of apoptosis. The results from the cell scratch test and Transwell test showed that emodin can significantly inhibit tumour migration. Apoptosis, also known as programmed cell death, is the basic mechanism that maintains the dynamic balance of the number of cells in the body. The suppression of apoptosis is an important cause of melanoma development. Apoptosis is mainly triggered by four pathways: the mitochondrial pathway, death receptor-mediated pathway, endoplasmic reticulum pathway and caspase-independent pathway. Caspase-3 is a protein shared in the first two pathways. It is a key factor that executes apoptosis, making it an important protein in the process of apoptosis. The activation of the mitochondrial pathway or death receptor-mediated pathway can cause an increase in caspase-3 expression. Its expression level also reflects the extent of apoptosis. The TUNEL assay showed an increased number of fluorescently stained cells, suggesting that emodin promoted the apoptosis of the melanoma cells. Moreover, the results of the western blot analysis showed that the expression of caspase-3 increased in the cells treated with 40 and 60 µM emodin, supporting the hypothesis that emodin may cause the induction of apoptosis through the mitochondrial pathway or death receptor-mediated pathway.
Conclusions
In conclusion, the results showed that emodin effectively inhibited the proliferation of B16F10 cells, inhibited tumour cell migration and promoted cell apoptosis. Emodin may be one of the compounds for improving the therapeutic methods used for melanoma.
Acknowledgments
Funding: This work was supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1040
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1040
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (http://dx.doi.org/10.21037/tcr-20-1040). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee C, Collichio F, Ollila D, et al. Historical review of melanoma treatment and outcomes. Clin Dermatol 2013;31:141-7. [Crossref] [PubMed]
- Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. Br J Dermatol 2014;170:11-9. [Crossref] [PubMed]
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374-403. [Crossref] [PubMed]
- Bradford PT, Anderson WF, Purdue MP, et al. Rising melanoma incidence rates of the trunk among younger women in the United States. Cancer Epidemiol Biomarkers Prev 2010;19:2401-6. [Crossref] [PubMed]
- Linos E, Swetter SM, Cockburn MG, et al. Increasing burden of melanoma in the United States. J Invest Dermatol 2009;129:1666-74. [Crossref] [PubMed]
- Alisi A, Pastore A, Ceccarelli S, et al. Emodin Prevents Intrahepatic Fat Accumulation, Inflammation and Redox Status Imbalance During Diet-Induced Hepatosteatosis in Rats. Int J Mol Sci 2012;13:2276-89. [Crossref] [PubMed]
- Lee YS, Kang OH, Choi JG, et al. Synergistic effect of emodin in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. Pharm Biol 2010;48:1285-90. [Crossref] [PubMed]
- Lu Y, Yang JH, Li X, et al. Emodin, a naturally occurring anthraquinone derivative, suppresses IgE-mediated anaphylactic reaction and mast cell activation. Biochem Pharmacol 2011;82:1700-8. [Crossref] [PubMed]
- Fu ZY, Han JX, Huang HY. Effects of emodin on gene expression profile in small cell lung cancer NCI-H446 cells. Chin Med J (Engl) 2007;120:1710-5. [Crossref] [PubMed]
- Li WY, Chan RY, Yu PH, et al. Emodin induces cytotoxic effect in human breast carcinoma MCF-7 cell through modulating the expression of apoptosis-related genes. Pharm Biol 2013;51:1175-81. [Crossref] [PubMed]
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol 2012;226:352-64. [Crossref] [PubMed]
- Weber J, Mandala M, Del Vecchio M, et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N Engl J Med 2017;377:1824-35. [Crossref] [PubMed]
- Maio M, Lewis K, Demidov L, et al. Adjuvant vemurafenib in resected, BRAF(V600) mutation-positive melanoma (BRIM8): a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol 2018;19:510-20. [Crossref] [PubMed]
- Huang Q, Lu G, Shen HM, et al. Anti-cancer properties of anthraquinones from rhubarb. Med Res Rev 2007;27:609-30. [Crossref] [PubMed]
- Yu JQ, Bao W, Lei JC. Emodin regulates apoptotic pathway in human liver cancer cells. Phytother Res 2013;27:251-7. [Crossref] [PubMed]
- Jiang J, Zhou N, Ying P, et al. Emodin promotes apoptosis of human endometrial cancer through regulating the MAPK and PI3K/ AKT pathways. Open Life Sci 2019;13:489-96. [Crossref]
- Liu H, Xu H, Zhang C, et al. Emodin-Loaded PLGA-TPGS Nanoparticles Combined with Heparin Sodium-Loaded PLGA-TPGS Nanoparticles to Enhance Chemotherapeutic Efficacy Against Liver Cancer. Pharm Res 2016;33:2828-43. [Crossref] [PubMed]
- Tabolacci C, Lentini A, Mattioli P, et al. Antitumor properties of aloe-emodin and induction of transglutaminase 2 activity in B16-F10 melanoma cells. Life Sci 2010;87:316-24. [Crossref] [PubMed]
- Yaoxian W, Hui Y, Yunyan Z, et al. Emodin induces apoptosis of human cervical cancer hela cells via intrinsic mitochondrial and extrinsic death receptor pathway. Cancer Cell Int 2013;13:71. [Crossref] [PubMed]