
Discovered differentially expressed lncRNA AC010973.2 can act as a diagnostic and prognostic biomarker for colon adenocarcinoma
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed malignancy and one of the leading causes of cancer-related mortality globally (1). Colon adenocarcinoma (COAD) is a common type of CRC (2). Recent research on COAD has made significant progress, but the data show that the morbidity and mortality rates of COAD are increasing (3). Findings have demonstrated that COAD can be treated successfully when identified at an early stage (4). The disease can cause blood in the stool, stomach pain, and a change in bowel movements, but some people remain asymptomatic, which poses a challenge for early diagnosis (5). Biomarkers currently play an essential role in the detection and treatment of patients with COAD (6).
Long noncoding RNAs (lncRNAs) are noncoding transcripts, usually longer than 200 nucleotides, that have recently emerged as one of the largest and significantly diverse RNA families (7). LncRNAs modulate various biological functions at the epigenetic (8), transcriptional (9), and post-transcriptional (10) levels, or directly regulate protein activity (11). The dysregulation of lncRNA expression can also lead to various diseases, including diabetes (12), obesity (13), osteoporosis (14), and various cancers (15,16). Nevertheless, current knowledge concerning lncRNA regulation in COAD is limited, requiring further exploration and accumulation of more evidence (17).
In this study, we downloaded the COAD transcript sequencing data from The Cancer Genome Atlas (TCGA). After analysis to obtain differentially expressed lncRNA, a series of bioinformatics analyses was carried out on these differentially expressed lncRNAs to construct a COAD diagnostic model. Based on the results, we determined that lncRNA AC010973.2 can be used as a reliable prognostic marker for COAD. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2011).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Public data acquisition and re-annotation
The gene expression quantification and corresponding clinical information of COAD patients were obtained from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). The obtained population comprised of 480 COAD tumor tissue samples and 41 adjacent non-tumor tissue samples (from 41 COAD patients). Both the mRNA expression data and the clinicopathologic characteristics of COAD are publicly available on TCGA.
After obtaining the raw data of COAD from TCGA, we re-annotated it using gene transfer format (GTF) (ftp://ftp.ensembl.org/pub/release-89/gtf/homo_sapiens/) and noted mRNA properties (coded and non-coded). We obtained the lncRNA expression values by repurposing the probes in the mRNA expression profiles to lncRNA. Next, the differentially expressed lncRNAs were analyzed by the R/Bioconductor package of edgeR (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) (18). Differentially expressed lncRNAs in the data set with |log2fold change (FC)| ≥1 and P<0.05 were included in subsequent analyses.
LncRNA risk model construction
After obtaining the survival time and survival status information of the COAD patients from TCGA, the lncRNA expression data and the clinical data were integrated through the patient ID, using Perl Language. Death was marked as 1 and survival as 0. Univariate Cox models were used to determine the association between the expression level of lncRNAs and the patient’s overall survival (OS). Differences with P<0.05 were considered statistically significant. We used the survival package of R to conduct a univariate cox analysis on the above integrated data to screen for lncRNAs associated with survival, with P<0.001 as the standard. We also used the survminer package of R to perform a multivariate analysis. According to the risk value obtained for each patient from the multivariate analysis, we calculated the median risk value for these patients. Based on this, we divided the patients into high-risk and low-risk groups. We then calculate the survival difference between the groups, using a survival curve and a forest map according to the hazard ratio. To display the survival risk data and the status of each patient more intuitively, we plotted the risk heatmap, risk curve, and survival status chart according to the patient’s risk value. Finally, we constructed a COAD-lncRNA risk model (Cox model).
Assessment of the Cox model
The 5-year-dependent receiver operating characteristic (ROC) curve analysis was performed to estimate the patient survival predictive accuracy of the Cox model. The performance of the Cox model was evaluated by the area under the ROC curve (AUC). We also calculated the concordance index (C-index) using the survcomp package of R to further evaluate the prediction accuracy of the Cox model.
Prediction of lncRNA-related protein-coding genes (PCGs)
We examined the correlation between the expression level of the lncRNAs and each PCG using two-sided Pearson correlation coefficients and the Z-test. PCGs considered as lncRNA-related if they were positively or negatively correlated with these lncRNAs. |Pearson correlation coefficient| >0.4 and P<0.001 were used to indicate significant correlation.
Function and pathway enrichment analysis
Through the DOSE package of R/Bioconductor, and the clusterProfiler, pathview, and org.Hs.eg.db packages of R, we conducted an enrichment analysis of lncRNAs in the Cox model. Our pathway enrichment analyses included mainly tools from the Gene Ontology (GO) (19) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (20).
Validation of lncRNA-related PCGs expression levels
The expression levels of lncRNA-related PCGs in COAD and normal samples were verified by Gene Expression Profiling Interactive Analysis (GEPIA). This is a web-based tool that delivers fast and customizable functionalities based on TCGA and Genotype Tissue Expression (GTEx) data. GEPIA has key interactive and customizable functions, including differential expression analysis, profiling plotting, correlation analysis, patient survival analysis, similar gene detection, and dimensionality reduction analysis (21). Hub genes with |log2FC| >1 and P<0.05 were considered as statistically significant.
Validation of lncRNA-related PCGs survival
The survival of lncRNA-related PCGs in COAD patients was verified by GEPIA. The survival results are presented as diagrams. The hazard ratio was calculated based on the Cox proportional-hazards (PH) model; cutoff-high and cutoff-low were both 50%.
Prediction of miRNA targets for lncRNA
To further explore the potential role of lncRNA in colon cancer, we used the DIANA-LncBase v3 (22) online database to predict the miRNA targets of lncRNA. This database (www.microrna.gr/LncBase) is a reference repository with experimentally supported miRNA targets on non-coding transcripts.
Statistical analysis
Most of the statistical analyses were performed using the bioinformatic tools mentioned above. When we conducted differential expression analysis, only lncRNA with |log2FC| >1 and P<0.05 were considered as statistically significant. Cox P<0.05 was regarded as statistically significant for survival analysis.
Results
Differentially expressed lncRNA profiles
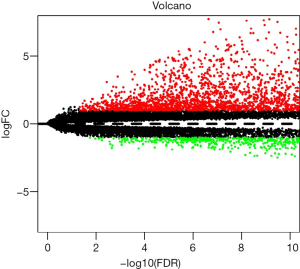
Through re-annotation and differential analysis of 20,406 mRNAs in TCGA data, we obtained 4,551 differentially expressed coding mRNAs and 1,679 differentially expressed lncRNAs between COAD patients and normal samples (online table, available at: https://cdn.amegroups.cn/static/application/421c784a070eeb20dc2474925b6f9ca8/10.21037tcr-20-2011-ts1.pdf). These differentially expressed lncRNAs include 283 upregulated lncRNAs and 1,396 downregulated lncRNAs (Figure 1).
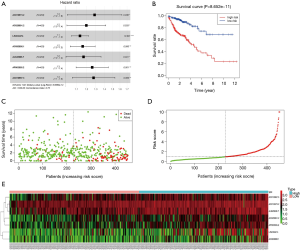
Establishment of the lncRNA risk model
We obtained 158 survival-related lncRNAs through a univariate Cox survival analysis of the 1,679 differentially expressed lncRNAs (online table, available at: https://cdn.amegroups.cn/static/application/3814a66cb3bb5fd776359343b3662770/10.21037tcr-20-2011-ts2.pdf). These lncRNAs were screened with P<0.001 as the standard, resulting in the following 11 lncRNAs that were survival-related at a very high significance level: AP003555.2, AC093895.1, AC010973.2, LINC02474, AC133528.1, AC010997.3, AC020891.2, AL645608.7, AP006284.1, and LINC02257. Through multivariate Cox analysis of these 11 lncRNAs, we narrowed the list to the following seven survival-related lncRNAs: AC010973.2, AC020891.2, LINC02474, AC093895.1, AL645608.7, AP003555.2, and AC010997.3 (Figure 2A). According to the expression level and risk coefficient of these seven lncRNAs, we constructed a COAD-lncRNA risk model. We also plotted survival curves, risk score, survival status, and expression of lncRNAs in COAD patients (Figure 2B,C,D,E). The clinical information of all COAD patients (including age, gender, survival status and cancer stage) is organized in the online table (available at: https://cdn.amegroups.cn/static/application/29574cb264fcfea3e286bb2d87f3ec4e/10.21037tcr-20-2011-ts3.pdf).
Assessment of the Cox model
By analyzing the 5-year survival data, we obtained the ROC curve, with an AUC =0.758, indicating that the Cox model has good accuracy. A C-index =0.753 further supports the accurate prediction of the Cox model (Figure 3 and Table 1).
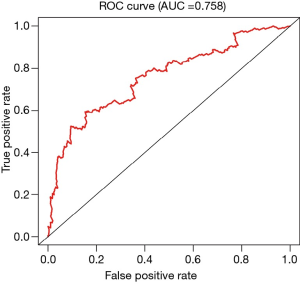
Table 1
| C-index | SE (SD) | Lower | Upper | P value |
|---|---|---|---|---|
| 0.7527377 | 0.02473711 | 0.7042539 | 0.8012216 | 1.665076e-24 |
The PCGs related to the lncRNAs
By examining the correlation between the expression level of the seven lncRNAs and the PCGs using two-sided Pearson’s correlation coefficients and the Z-test, we obtained a list of PCGs that are related to these seven lncRNAs (online table, available at: https://cdn.amegroups.cn/static/application/b8aaf6ceb0eeac9699c2750e5562faba/10.21037tcr-20-2011-ts4.pdf). We identified 1,457 AC010973.2-related PCGs (including ZNF692, HSF4, CDK10, FAM160B1), 28 AC020891.2-related PCGs, 2 LINC02474-related PCGs, 7 AC093895.1-related PCGs, 16 AL645608.7-related PCGs, 6 AP003555.2-related PCGs, and 97 AC010997.3-related PCGs.
GO and KEGG enrichment analyses
The GO enrichment analysis of the 1,457 AC010973.2-related PCGs indicated that this lncRNA is associated with activities related to nucleoside-triphosphatase regulator, GTPase regulator, GTPase activator, protein serine/threonine kinase, ATP-dependent helicase, pure nucleotide triphosphate (NTP)-dependent helicase, SH3 domain binding catalysis, acting on RNA alpha-mannosidase, phosphatase regulator, catalysis, and DNA-related (Figure 4A,B and online table, available at: https://cdn.amegroups.cn/static/application/f5dba0e31a79fb91ee61fc0c97debf51/10.21037tcr-20-2011-ts5.pdf).
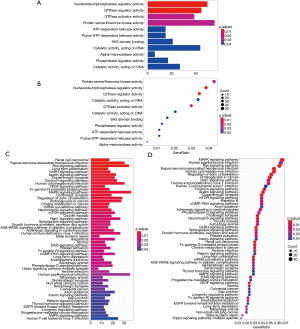
The result of the KEGG enrichment analysis of these 1,457 AC010973.2-related PCGs indicate that AC010973.2 has a significant correlation with 55 pathways, including CRC (READ), choline metabolism in cancer, and the AMPK signaling pathway (Figure 4C,D and online table, available at: https://cdn.amegroups.cn/static/application/f5dba0e31a79fb91ee61fc0c97debf51/10.21037tcr-20-2011-ts5.pdf).
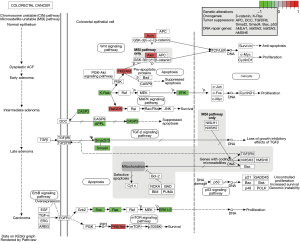
In the CRC pathway (Figure 5), AC010973.2-related PCGs included SOS2, RALGDS, AKT2, SOS1, KRAS, AXIN1, HRAS, NRAS, CASP3, MAPK1, SMAD2, SMAD4, and APPL1. We plotted the co-expression maps of AC010973.2 and 13 CRC pathways obtained based on the AC010973.2-related PCGs (Figure 6). AC010973.2 positively-related PCGs include RALGDS, AKT2, AXIN1, HRAS, MAPK1, SMAD2, SMAD4, and APPL1. AC010973.2 negatively-related PCGs include SOS2, SOS1, KRAS, NRAS, and CASP3.
Expression and survival analyses
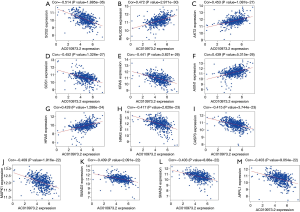
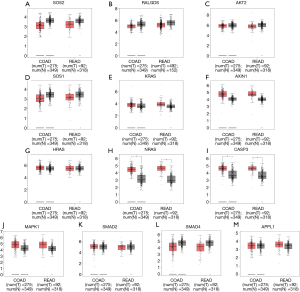
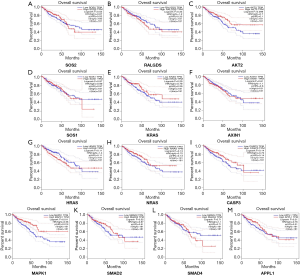
To further evaluate the 13 CRC pathways associated with the AC010973.2-related PCGs, we performed expression and survival analyses through the GEPIA database, and the results are shown in Figures 7,8. NRAS and CASP3 were markedly upregulated in COAD/READ tissues when compared with normal tissues. Patients whose samples showed high expression of MAPK1 had a higher survival rate than those whose samples had low expression levels, suggesting a batter prognostic value for COAD/READ.
LncRNA target miRNA prediction
To further explore the potential mechanism of action of lncRNA AC010973.2, we used DIANA-LncBase v3 to predict its target miRNA. A total of 20 miRNAs are predicted, including miR-101-3p, miR-1304-3p, miR-15a-3p, miR-191-5p, miR-193a-3p, miR-193b-3p, miR-200c-3p, miR-204-5p, miR-210-3p, miR-219a-1-3p, miR-22-3p, miR-27b-5p, miR-29c-5p, miR-301a-5p, miR-3127-3p, miR-3176, miR-4326, miR-500a-5p, miR-550a-5p and miR-7-5p (the mechanism of these miRNAs in CRC is summarized in online table, available at: https://cdn.amegroups.cn/static/application/086702039c8bad7c9b5231bf5476298e/10.21037tcr-20-2011-ts6.pdf).
Discussion
Effective management of COAD depends on early diagnosis and proper monitoring of response to therapy. However, these goals are difficult to achieve because of the lack of sensitive and specific biomarkers for early detection and disease monitoring (23). By mining TCGA COAD expression data, using differential, and univariate and multivariate Cox analyses, we constructed a COAD-Cox model. In this model, seven lncRNAs are positively associated with the risk level in COAD. After the model was successfully constructed, we used AUC of ROC analysis and C-index to evaluate the accuracy of the model. Both AUC (0.758) and C-index (0.753) were greater than 0.7, indicating that our model is accurate and can be used to evaluate the patients’ risk.
The enrichment analyses using GO and KEGG databases showed that lncRNA AC010973.2 is related to the mitogen-activated protein kinase (MAPK) signaling pathway, READ pathway, and others. The MAPK cascades are key signaling pathways that regulate a wide variety of cellular processes, including proliferation, differentiation, apoptosis, and stress responses (24). Several studies have shown that the MAPK signaling pathway is closely related to cancer progression (25-28). A few studies have shown that lncRNA can regulate cancer through the MAPK signaling pathway (29-31). Our analysis shows that lncRNA might be associated with the MAPK signaling pathway and CRC, which implies that lncRNA may affect the occurrence of CRC through the MAPK signaling pathway.
Through expression and survival analyses of 13 lncRNA-related PCGs, we found that the AC010973.2 negatively-related PCGs NRAS and CASP3 have higher expression in COAD/READ patients, while the high expression level of the AC010973.2 positively-related PCG MAPK1 correlated with a high survival rate of COAD/READ patients. This implies that AC010973.2 plays a protective role against the progression of COAD/READ. This is, however, contrary to what the multivariate Cox model suggests. Based on the model, AC010973.2 is a high-risk factor. We suspect that such a situation occurs because AC010973.2 does not directly interact with these genes, but functions through other genes or pathways.
MiRNAs are small non-coding RNAs that function as guide molecules in RNA silencing (32). MiRNAs are involved in nearly all developmental and pathological processes in animals by targeting most protein-coding transcripts (33,34). Misregulation of miRNA expression can cause many diseases, including cancer (35). The competing endogenous RNA (ceRNA) hypothesis links lncRNA and miRNA into a large-scale regulatory network across the transcriptome. This network greatly expands the functional genetic information in the human genome and plays an important role in pathological conditions such as cancer (36). A number of studies have shown that lncRNA acts as a miRNA sponge. Through this action, lncRNA affects the occurrence of cancer by affecting the binding of miRNA to target genes (36-38). We predict that lncRNA AC010973.2 might be acting as a sponge for some miRNAs and, by doing so, affects the occurrence of COAD.
AC010973.2 is an antisense lncRNA. Its specific genomic location is chr7: 151074742–151076530, it is 1,230 bp long (39), and about half of it is located in the cytoplasm (40). SLC4A2 is the homologous host gene of AC010973.2. It was shown that disruption of SLC4A2 is related to the occurrence of COAD (41). AC010973.2 might be affecting the occurrence of COAD by regulating the expression of SLC4A2. A literature search produced no reports on the function of AC010973.2. We can, therefore, state that through bioinformatics, we have found a new lncRNA that is significantly related to COAD survival. This finding provides new directions for the diagnosis and treatment of COAD.
Although we found data on a large number of COAD samples through TCGA and conducted detailed bioinformatics analysis on them, the current work is still insufficient. All our conclusions are based on computational analyses, without any verification tests in vivo or in vitro. Therefore, studying the function of AC010973.2 in in vivo and in vitro experiments will be an important part of our future work.
Conclusions
In conclusion, using a multivariate Cox analysis model, we identified lncRNA AC010973.2, whose expression profile is significantly related to the survival of COAD patients. Our results suggest that AC010973.2 might be a useful biomarker for prognosis, leading to an increasingly personalized therapeutic approach. The unknown function of AC010973.2, and the homologous host gene that is significantly related to COAD, open the way to a new direction for further research on the occurrence and prognosis of COAD.
Acknowledgments
Funding: This research was funded by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2011
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2011). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:1298-306. [Crossref] [PubMed]
- Cai L, Bennedsen ALB, Qvortrup C, et al. Increasing incidence of colorectal cancer in young patients. Ugeskr Laeger 2019;182:V09190524. [PubMed]
- Roncucci L, Mariani F. Prevention of colorectal cancer: How many tools do we have in our basket? Eur J Intern Med 2015;26:752-6. [Crossref] [PubMed]
- Wilkins T, Reynolds PL. Information from your family doctor: colon cancer screening. Am Fam Physician 2008;78:1393-4. [PubMed]
- Lech G, Słotwiński R, Słodkowski M, et al. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol 2016;22:1745-55. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol 2016;1402:271-86. [Crossref] [PubMed]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science 2012;338:1435-9. [Crossref] [PubMed]
- Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem 2017;292:12375-82. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Liu CY, Zhang YH, Li RB, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun 2018;9:29. [Crossref] [PubMed]
- Chang W, Wang J. Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells. 2019;8:853. [Crossref] [PubMed]
- Dallner OS, Marinis JM, Lu YH, et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat Med 2019;25:507-16. [Crossref] [PubMed]
- Mei B, Wang Y, Ye W, et al. LncRNA ZBTB40-IT1 modulated by osteoporosis GWAS risk SNPs suppresses osteogenesis. Hum Genet 2019;138:151-66. [Crossref] [PubMed]
- Wang Y, Wang Z, Xu J, et al. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat Commun 2018;9:3192. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Xing Y, Zhao Z, Zhu Y, et al. Comprehensive analysis of differential expression profiles of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic signature for patients with colon adenocarcinoma. Oncol Rep 2018;39:2365-75. [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res 2004;32:D277-80. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Karagkouni D, Paraskevopoulou MD, Tastsoglou S, et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res 2020;48:D101-10. [PubMed]
- Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res 2016;36:1093-102. [PubMed]
- Guo YJ, Pan WW, Liu SB, et al. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 2020;19:1997-2007. [PubMed]
- Xu W, Gu J, Ren Q, et al. NFATC1 promotes cell growth and tumorigenesis in ovarian cancer up-regulating c-Myc through ERK1/2/p38 MAPK signal pathway. Tumour Biol 2016;37:4493-500. [Crossref] [PubMed]
- Qi K, Li Y, Li X, et al. Id4 promotes cisplatin resistance in lung cancer through the p38 MAPK pathway. Anticancer Drugs 2016;27:970-8. [Crossref] [PubMed]
- Zhao D, Zhang T, Hou XM, et al. Knockdown of fascin-1 expression suppresses cell migration and invasion of non-small cell lung cancer by regulating the MAPK pathway. Biochem Biophys Res Commun 2018;497:694-9. [Crossref] [PubMed]
- Peluso I, Yarla NS, Ambra R, et al. MAPK signalling pathway in cancers: Olive products as cancer preventive and therapeutic agents. Semin Cancer Biol 2019;56:185-95. [Crossref] [PubMed]
- Tasharrofi B, Ghafouri-Fard S. Long Non-coding RNAs as Regulators of the Mitogen-activated Protein Kinase (MAPK) Pathway in Cancer. Klin Onkol 2018;31:95-102. [Crossref] [PubMed]
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:443-9. [PubMed]
- Qu CX, Shi XC, Bi H, et al. LncRNA AOC4P affects biological behavior of gastric cancer cells through MAPK signaling pathway. Eur Rev Med Pharmacol Sci 2019;23:8852-60. [PubMed]
- Catalanotto C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int J Mol Sci 2016;17:1712. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019;20:5-20. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Du W, Feng Z, Sun Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem Biophys Res Commun 2018;507:198-202. [Crossref] [PubMed]
- Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis 2018;9:982. [Crossref] [PubMed]
- Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982. [Crossref] [PubMed]
- Volders PJ, Anckaert J, Verheggen K, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res 2019;47:D135-9. [Crossref] [PubMed]
- Cao Z, Pan X, Yang Y, et al. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 2018;34:2185-94. [Crossref] [PubMed]
- Hulikova A, Black N, Hsia LT, et al. Stromal uptake and transmission of acid is a pathway for venting cancer cell-generated acid. Proc Natl Acad Sci U S A 2016;113:E5344-53. [Crossref] [PubMed]


