
High expression of forkhead box M1 (FOXM1) is a poor prognostic biomarker in lung adenocarcinoma
Introduction
Lung cancer is one of the most common causes of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for 80% to 85% of all lung cancer cases, and lung adenocarcinoma (ADC) and lung squamous cell carcinoma (SCC) are the main histopathologic subtypes of NSCLC (1,2). ADC and SCC are very similar in clinical manifestations, but have significant differences in pathogenesis, treatment and prognosis (3). For example, activating mutations in the epidermal growth factor receptor (EGFR) and in the anaplastic lymphoma kinase (ALK) fusion typically occur in lung ADC but not lung squamous cell cancer, rendering drugs targeting these genes ineffective against lung squamous cell cancer (4). Further study of these differences in subtype characteristics will contribute to a deeper understanding and identification of lung cancers and molecular targeting strategies for treatment.
Forkhead box M1 (FOXM1) is one of the important members of the forkhead box (FOX) transcription factor family, whose members are characterized by a DNA-binding domain (DBD), also referred to as a forkhead or winged helix domain, consisting of greater than 100 amino acids (5). FOXM1 is a proliferation-related transcription factor whose expression is increased in proliferating cells, but decreased or absent in cells undergoing terminal differentiation (6). FOXM1 is also highly expressed in embryos, whereas in adults, it is only expressed in self-renewing tissues, such as thymus and testis, and a few proliferating cells (7). FOXM1 is a crucial regulator of the cell cycle (8), and mainly promotes G1-S and G2-M transitions during cell cycle progression, and ensures mitotic spindle integrity by positively regulating transcription of its downstream cell cycle-specific genes (9,10). FOXM1 is also involved in various of biological processes, such as cell proliferation, apoptosis, differentiation, angiogenesis, and DNA damage repair (11). Normal expression and activity maintain the balance of transcriptional programs, ensuring appropriate growth and maturation during the development of embryos and fetuses, as well as maintaining proper homeostasis and normal repair of adult tissues. On the contrary, abnormal expression and activity of FOXM1 may cause the biological processes in which it is involved to be disordered, leading to tumor formation, progression, invasion and metastasis (12). Previous studies have found that FOXM1 expression is up-regulated in lung cancer and leads to poor prognosis. However, the expression and prognostic value for different pathological types of lung cancer have not been further studied (13,14). Moreover, the mechanisms of FOXM1 mRNA expression dysregulation are far from being fully understood. Therefore, we investigate FOXM1 mRNA expression and its clinical value for patients with ADC and SCC.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-1103).
Methods
Oncomine analysis
The Oncomine database version 4.5 (https://www.oncomine.org/) was used to characterize FOXM1 profiles in lung cancer (15). We compared FOXM1 mRNA levels in lung cancer tissues vs. normal tissues in different datasets using the following threshold: fold change ≥2, P<0.05, gene rank: top 10%.
TCGA and cBioPortal analysis
The Cancer Genome Atlas (TCGA) data from the cBioPortal database (16,17) (http://www.cbioportal.org/) was used to identify the association between FOXM1 mRNA expression, clinical variables and prognosis, as well as copy number alterations (CNAs) and DNA methylation in ADC vs. SCC patients. FOXM1 mRNA expression z-scores (RNA Seq V2 RSEM), DNA methylation (HM450), copy-number alterations (GISTIC) and survival data, including OS status, OS time, recurrence status and recurrence-free survival (RFS) time, were downloaded from the cBioPortal TCGA lung ADC cohort (n=586) (TCGA, provisional) and TCGA lung SCC cohort (n=511) (TCGA, provisional).
FOXM1 mRNA levels were divided into two groups (high expression and low expression) by setting the median expression as the cutoff. CNAs were defined in each gene by sample-specific thresholds as follows: deep deletion (−2), shallow deletion (−1), neutral/diploid (0), gain (+1) and amplification (+2) by using the Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm. Patients were divided into 3 groups: a FOXM1 copy number amplification group (positive values), copy number loss group (negative values) and normal group (zero). For grouping DNA methylation, we calculated the 75th percentile of β-values from all analyzed patients and used it as the cut-off value. Subsequently, individuals with a β-value higher than the cut-off value were considered methylated, otherwise individuals were considered unmethylated.
Co-expression and network analysis
We used the cBioPortal database online tool (http://www.cbioportal.org/) (16,17) to construct a network for FOXM1 and the significantly altered neighboring genes in ADC (TCGA, Provisional). Gene Ontology (GO) enrichment analysis of the enriched biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed by using the DAVID Bioinformatics Resources 6.8 online platform (https://david-d.ncifcrf.gov/home.jsp) (18,19). Enrichment results with both a P value and false discovery rate (FDR) <0.05 were considered statistically significant.
Statistical analysis
Data were analyzed with SPSS 24.0 software (SPSS Inc.) and graphical data were constructed using GraphPad Prism 6.0 software (GraphPad Inc.). We used a two-tailed Student’s t-test to identify the association between FOXM1 mRNA expression and clinical pathological features (age, sex, T stage, N stage, M stage, TNM stage and smoking history) of ADC patients or SCC patients. We used Kaplan-Meier curves and the log-rank test to analyze the correlation between FOXM1 mRNA expression and OS or RFS of ADC patients and SCC patients. We used Cox regression analysis to estimate whether FOXM1 mRNA expression was an independent risk factor for OS or ADC patients. The correlation between FOXM1 mRNA expression and DNA CNAs or DNA methylation was conducted by the Spearman or Pearson test. A P value <0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
FOXM1 mRNA expression is elevated in lung cancer tissues
We investigated FOXM1 mRNA expression in lung cancer by using the Oncomine database. Results showed that expression of FOXM1 mRNA was increased in lung cancer tissues compared to normal tissues (Figure 1A). Two large sample size studies, by Bhattacharjee et al. (Figure 1B) and Hou et al. (Figure 1C), revealed that FOXM1 mRNA expression was specifically elevated in the lung cancer group (P=0.0026 and P<0.0001, respectively). There was a total of 14 studies, including 783 samples and fold changes between 3.167 and 44.293, and P values from 4.00E-03 to 7.67E-30, showing elevated FOXM1 in lung cancer (Table 1).

Table 1
| Study name | Cancer type | Normal, n | Cancer, n | Total, n | Fold change | P value |
|---|---|---|---|---|---|---|
| Hou Lung | Squamous cell lung carcinoma | 65 | 27 | 92 | 9.019 | 7.67E-30 |
| Hou Lung | Lung adenocarcinoma | 65 | 45 | 110 | 3.768 | 1.05E-13 |
| Hou Lung | Large cell lung carcinoma | 65 | 19 | 84 | 8.045 | 3.23E-07 |
| Bhattacharjee Lung | Squamous cell lung carcinoma | 17 | 21 | 38 | 44.293 | 5.53E-09 |
| Bhattacharjee Lung | Small cell lung carcinoma | 17 | 6 | 23 | 15.129 | 2.63E-06 |
| Bhattacharjee Lung | Lung adenocarcinoma | 17 | 132 | 149 | 3.257 | 4.00E-03 |
| Garber Lung | Small cell lung carcinoma | 6 | 4 | 10 | 5.209 | 2.58E-05 |
| Garber Lung | Squamous cell lung carcinoma | 6 | 13 | 19 | 4.105 | 2.73E-06 |
| Garber Lung | Lung adenocarcinoma | 6 | 40 | 46 | 3.339 | 3.83E-06 |
| Garber Lung | Large cell lung carcinoma | 6 | 4 | 10 | 7.252 | 6.40E-04 |
| Su Lung | Lung adenocarcinoma | 30 | 27 | 57 | 4.025 | 2.05E-09 |
| Beer Lung | Lung adenocarcinoma | 10 | 86 | 96 | 26.543 | 2.07E-07 |
| Stearman Lung | Lung adenocarcinoma | 19 | 20 | 39 | 4.725 | 1.01E-07 |
| Wachi Lung | Squamous cell lung carcinoma | 5 | 5 | 10 | 3.167 | 3.00E-03 |
Association between FOXM1 mRNA expression and clinicopathological parameters
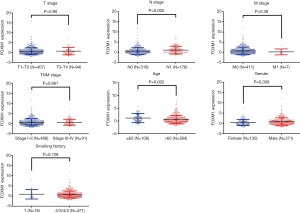
Because FOXM1 mRNA expression levels were higher in lung cancer, we further explored the associations between its mRNA expression and different clinicopathologic features in NSCLC patients via the TCGA database. We downloaded 586 ADC cases and 511 SCC cases, from the cBioPortal platform, and first compared the expression differences of FOXM1 mRNA in ADC and SCC patients. FOXM1 mRNA was more highly expressed in SCC patients than in ADC patients (P<0.0001, Figure 2). We then analyzed the relationship between FOXM1 mRNA expression and other clinical variables in ADC and SCC. Results showed that high expression of FOXM1 mRNA was correlated with higher N stage (P=0.02), advanced TNM stage (P=0.002), younger age at diagnosis (P=0.002), male patients (P=0.002) and longer smoking history (P=0.007) for ADC patients, but not with the T (P=0.3) or M stage (P=0.05) (Figure 3 and Table 2). However, in SCC patients, we observed that high expression of FOXM1 mRNA was only correlated with higher N stage (P=0.005), younger age at time of diagnosis (P=0.002) and male patients (P=0.009), but not with T stage (P=0.95), M stage (P=0.38), TNM stage (P=0.867) or smoking history (P=0.728) (Figure 4 and Table 2).
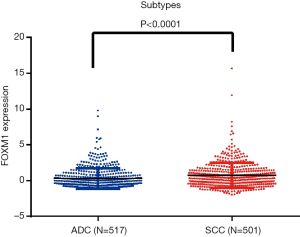
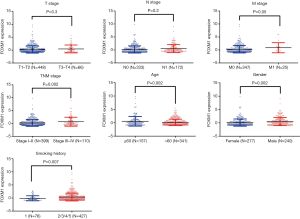
Table 2
| Variables | Classification | ADC (FOXM1 mRNA expression) | SCC (FOXM1 mRNA expression) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Mean | 95% CI | P value | Samples | Mean | 95% CI | P value | |||
| Gender | Female | 277 | 0.1073 | [0.1421, 0.6337] | 0.002* | 130 | 0.4063 | [0.1153, 0.8141] | 0.009* | |
| Male | 240 | 0.4952 | 371 | 0.8709 | ||||||
| Age | ≤60 | 157 | 0.5645 | [−0.6909, −0.1551] | 0.002* | 108 | 1.159 | [−0.9082, −0.216] | 0.002* | |
| >60 | 341 | 0.1416 | 384 | 0.5964 | ||||||
| T stage | T1-T2 | 448 | 0.2639 | [−0.1761, 0.566] | 0.302 | 407 | 0.7482 | [−0.3835, 0.4066] | 0.954 | |
| T3-T4 | 66 | 0.4588 | 94 | 0.7597 | ||||||
| N stage | N0 | 333 | 0.1992 | [0.4431, 0.5729] | 0.022* | 319 | 0.5911 | [0.1461, 0.7914] | 0.005* | |
| N1 | 172 | 0.5078 | 176 | 1.06 | ||||||
| M stage | M0 | 347 | 0.2812 | [−0.0026, 1.184] | 0.051 | 411 | 0.7702 | [−1.963, 0.7462] | 0.378 | |
| M1 | 25 | 0.8717 | 7 | 0.162 | ||||||
| TNM Stage | I-II | 399 | 0.1958 | [0.1678, 0.7708] | 0.002* | 406 | 0.7481 | [−0.4341, 0.3659] | 0.867 | |
| III-IV | 110 | 0.6651 | 91 | 0.714 | ||||||
| Smoking history | 1 | 76 | −0.1133 | [0.1322, 0.8301] | 0.007* | 18 | 0.8982 | [−0.9831, 0.6873] | 0.728 | |
| 2/3/4/5 | 427 | 0.3678 | 471 | 0.7503 | ||||||
Student’s t-test was used for statistical analysis. CI, confidence interval; T, the anatomical size of the primary tumors; N, the extent of lymph node involvement; M, the presence or absence of metastasis; TNM, tumor, node, metastasis; ADC, lung adenocarcinoma; SCC, lung squamous cell carcinoma. *, P<0.05 was considered statistically significant.
Prognostic value of increased FOXM1 mRNA expression in ADC and SCC patients
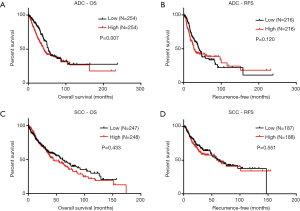
Because FOXM1 mRNA expression was significantly different between ADC and SCC patients, we examined whether this difference would lead to a difference in prognosis for patients of the two subtypes. To this end, we further explored the association between FOXM1 mRNA expression and patient prognosis in ADC and SCC patients. We used the Kaplan-Meier method to analyze the TCGA-ADC and TCGA-SCC survival data for OS and RFS. Results showed that ADC patients with high expression of FOXM1 mRNA had shorter OS (P=0.007, Figure 5A), but there was no statistical difference for RFS (P=0.12, Figure 5B). Surprisingly, we did not find any statistical differences for high or low FOXM1 mRNA in OS or RFS in SCC patients (P=0.433 and 0.551, respectively, Figure 5C,D).
To investigate the independent prognostic value of FOXM1 mRNA expression in ADC patients, we further conducted univariate and multivariate analyses of OS via TCGA data mining. Results demonstrated that, after adjustment for T stage, N stage, M stage and TNM stage, high FOXM1 mRNA expression was an independent prognostic factor for poor OS (P=0.012, HR, 1.574; 95% CI, 1.107 to 2.239, Table 3).
Table 3
| Variables | Classification | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | |||
| Age | >60 vs. ≤60 | 0.363 | 1.160 | [0.843, 1.596] | ||||
| Gender | Female vs. male | 0.715 | 1.055 | [0.790, 1.410] | ||||
| Smoking history | 2/3/4/5 vs. 1 | 0.722 | 0.928 | [0.615, 1.401] | ||||
| T Stage | T3-T4 vs. T1-T2 | 0.000* | 2.430 | [1.666, 3.543] | 0.004* | 1.971 | [1.235, 3.146] | |
| N Stage | N1 vs. N0 | 0.000* | 2.592 | [1.931, 3.480] | 0.000* | 2.044 | [1.380, 3.025] | |
| M Stage | M1 vs. M0 | 0.005* | 2.176 | [1.271, 3.727] | 0.319 | 1.392 | [0.727, 2.667] | |
| TNM Stage | III/IV vs. I/II | 0.000* | 2.267 | [1.961, 3.640] | 0.257 | 1.331 | [0.812, 2.180] | |
| FOXM1 expression | High vs. Low | 0.004* | 1.537 | [1.146, 2.062] | 0.012* | 1.574 | [1.107, 2.239] | |
HR, hazard ratio; CI, confidence interval; T, the anatomical size of the primary tumors; N, the extent of lymph node involvement; M, the presence or absence of metastasis; TNM, tumor, node, metastasis; ADC, lung adenocarcinoma; OS, overall survival. *, P<0.05 was considered statistically significant.
FOXM1 mRNA expression is positively correlated with FOXM1 CNAs, but negatively correlated with DNA methylation
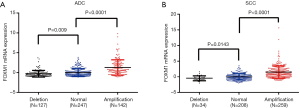
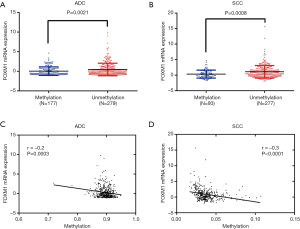
To investigate the possible mechanism responsible for the enhanced expression of FOXM1 mRNA in lung cancer, we analyzed the association between its mRNA expression and CNAs as well as DNA methylation by using the TCGA-ADC and TCGA-SCC data. There were 516 cases containing CNA data in ADC patients, and among these cases, 142 (27.6%) cases had FOXM1 copy number amplification (+1/+2). In SCC patients, there were 501 cases containing CNA data, including 259 (51.7%) cases with FOXM1 copy number amplification (+1/+2). We further calculated the association between FOXM1 mRNA expression and its CNAs. Findings showed that expression of FOXM1 mRNA was significantly elevated in the DNA copy number amplification group as compared to the normal DNA copy number group in both ADC and SCC patients (P<0.001, Figure 6A,B). Regression analysis confirmed a moderately positive correlation between FOXM1 mRNA expression and its CNAs (Spearman’s r=0.4 and 0.5 for ADC and SCC patients, respectively). We further examined the association between FOXM1 mRNA expression and DNA methylation. Results showed that compared with the unmethylated group, FOXM1 had lower mRNA expression in the methylated group (P=0.002 and P<0.0001 in ADC and SCC patients, respectively, Figure 7A,B). Moreover, regression analysis showed a weakly negative correlation between FOXM1 mRNA expression and DNA methylation (Pearson’s r=−0.2 and −0.3 in ADC and SCC patients, respectively, Figure 7C,D).
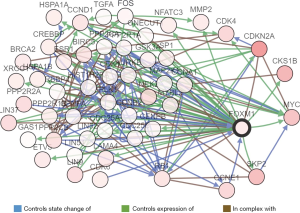
Predicted functions and pathway enrichment analysis of the frequently altered neighboring genes with FOXM1 in ADC patients
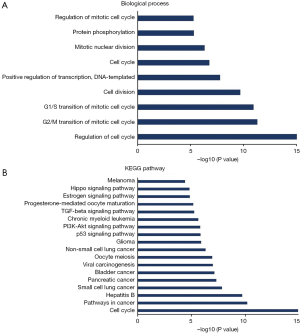
Duo to our results showed that high expression of FOXM1 mRNA is an independent prognostic indicator of OS in ADC patients, but not SCC patients, we predicted functions and pathways enrichment analysis to explore the possible regulatory mechanism of FOXM1 in ADC patients. A network for the top 50 altered neighboring genes with FOXM1 was constructed by the cBioPortal database to explain the mechanism of elevated FOXM1 mRNA expression resulting in worse OS in ADC patients. FOXM1 was closely associated with CCNE1, MYC, CKS1B, SKP2 and CDKN2A (Figure 8). Further GO functions and KEGG pathway enrichment analyses of these genes were performed using the DAVID online analysis tool. GO (Biological process) enrichment analysis showed that regulation of the cell cycle, involving G2/M and G1/S transitions, were associated with the FOXM1 alterations in ADC (Figure 9A). KEGG enrichment analysis found that there were 18 pathways related to the FOXM1 alterations in ADC (Figure 9B). Among these pathways, cell cycle, pathways in cancer, small cell lung cancer, NSCLC, p53 signaling pathway, PI3K-Akt signaling pathway and TGF-beta signaling pathway were related with tumorigenesis and progression of ADC.
Discussion
In our study, we used Oncomine database analysis to confirm that FOXM1 mRNA expression is increased in lung cancer. In particular, FOXM1 mRNA expression in SCC is distinctly higher than in ADC, especially in Bhattacharjee's study, which showed that the fold change was 44-fold increased in SCC, but only 3-fold increased in ADC. Moreover, we further show using the TCGA database, which has larger sample size, that FOXM1 mRNA expression is higher in SCC than in ADC.
Due to differences in the expression of FOXM1 at the mRNA level in ADC and SCC, we speculate that this difference may exert different regulatory effects on cancer cell behavior and may have different prognostic value in these two subtypes. First, we observed differences in FOXM1 mRNA expression and clinicopathological variables in ADC and SCC patients. In ADC patients, FOXM1 mRNA expression is correlated with N stage, TNM stage, age, gender and smoking history. However, in SCC patients, we observed that FOXM1 mRNA expression is only related to N stage, age and sex. Secondly, we further analyzed the association between FOXM1 mRNA expression and prognosis in ADC and SCC. Interestingly, although we found that FOXM1 mRNA is more highly expressed in SCC, Kaplan-Meier survival analysis did not find an association between FOXM1 mRNA expression and SCC patients for OS and RFS. However, our findings show that high expression of FOXM1 mRNA leads to worse OS in ADC patients, in whom we show, through multivariate analysis, that high expression of FOXM1 mRNA is an independent indicator of OS in ADC patients.
ADC and SCC have different features, such as location, cells of origin, pathology, oncogenic drivers, and molecular markers (20). Studies have reported that the two subtypes also have different prognoses. Han et al. suggested that SOX30 is a favorable and independent prognostic indicator in patients of ADC, but not in patients of SCC (21), and in another study showed that SOX30 binds to CTNNB1 to inhibit the Wnt/CTNNB1 signaling pathway, thereby suppressing tumor metastasis in early ADC, whereas in SCC, it does not bind to CTNNB1 and has no effect in SCC, which might be a reason result for the difference of prognosis in ADC and SCC patients (22). The results are similar to our study, which show that SCC consistently expresses a higher mRNA level of FOXM1 than ADC, but Kaplan-Meier analysis indicated that FOXM1 is a marker for poor OS in ADC patients, but not SCC patients. Therefore, we speculated that FOXM1 might have a similar regulatory network and mechanism to SOX30, which is worth further validating.
Both genetic and epigenetic alterations, such as CNAs and DNA methylation, can influence gene expression and modulate cancer cell behavior (23,24). We demonstrate a moderately positive and weakly negative association between FOXM1 mRNA expression and CNAs and DNA methylation, respectively. Findings suggest that FOXM1 CNA might be the main mechanism influencing its elevated mRNA expression in ADC and SCC patients. We also found that FOXM1 has a higher amplification ratio in SCC patients than in ADC patients (51.7% vs. 27.6%). This result is consistent with our result that FOXM1 mRNA expression is higher in SCC, which further supports the positive regulation of FOXM1 gene copy number on FOXM1 mRNA expression. However, in some samples with increased FOXM1 mRNA expression, there is no copy number amplification or lack of DNA methylation which indicates that other mechanisms are responsible for its upregulation. Recently, studies have focused on the correlation between mRNA expression and miRNA regulation. Ke et al. reported that miR-149 inhibited expression of FOXM1, thereby restoring epithelial-mesenchymal transition [EMT in NSCLC cells (25). Ma et al. also demonstrated that miR-509-5p exerts tumor-suppressive effects by attenuating FOXM1 in NSCLC (26). Therefore, down-regulation of miRNA may also be one of the reasons for up-regulation of FOXM1 in ADC and SCC.
To clarify the possible mechanism of FOXM1 mRNA high expression attributed to worse OS in ADC patients, we conducted a predictive network of the frequently altered neighboring genes with FOXM1 in ADC. Results showed that FOXM1 is associated with expression of MYC, SKP2 and CKS1B. Previously, a study indicated that FOXM1 regulates SKP2 expression and influences cell cycle G1-S transition (10). Zeng et al. also showed depletion of FOXM1 leads to the decreased expression of its targets SKP2 and c-MYC in gastric cancer cells (27). Therefore, we hypothesize that elevated FOXM1 mRNA expression in ADC leads to upregulation of MYC and SKP2, thereby promoting tumor cell proliferation. In addition, CCNE1 involved in the state change of FOXM1 One study indicated that Cyclin E binds to CDK2, then leads to phosphorylation of retinoblastoma-1 (Rb-1) or other substrates, which regulates cell cycle G1-S phase transition (28). Furthermore, another study has revealed that Cyclin E overexpression associated with poor prognosis in NSCLC (29). Whether CCNE1 also directly binds to and phosphorylates FOXM1, thus exerting carcinogenic effects to contribute to poor OS of ADC patients deserves further investigation. Furthermore, Hayashi et al. determined that cyclin E expression is lower in ADC than in SCC (30). This finding supports our hypothesis because we also found that FOXM1 has lower expression in ADC than in SCC. In addition, our results showed that FOXM1 and CDKN2A maybe be the same complex and interact with each other. Study indicated that CDKN2A is one of the transcriptional targets of FOXM1, which is also a critical gene for the NF-kB and JNK pathways. FOXM1 can induce its expression and then involve in KrasG12D-mediated lung tumorigenesis (31). Moreover, transgenic overexpression of activated KrasG12D in mouse respiratory epithelium can induce lung ADCs (32). These reports fully demonstrate that high FOXM1 mRNA expression may contribute to poor OS in ADC patients by acting on these key upstream and downstream molecules. GO function enrichment analysis showed that the altered neighboring genes with FOXM1 in ADC are mainly involved in regulating the cell cycle, including G2/M transition and G1/S transition. KEGG enrichment analysis found that FOXM1 could mediate the p53, PI3K-Akt and TGF-beta signaling pathways. These are common biological processes and pathways that have been identified to be closely related to tumorigenesis and progression (33). In our study of FOXM1 mRNA expression, the association between its expression and clinicopathologic parameters, as well as patient prognosis, the network of FOXM1 and its co-expressed genes, and the function and pathway enrichments were based on data mining of the Oncomine and TCGA databases. Further in vivo and in vitro experiments and clinical trials are required to verify our conclusions.
Conclusions
The present study demonstrates that although FOXM1 mRNA expression is higher in SCC than in ADC, it is associated with poor OS in patients with ADC, but not in patients with SCC. Therefore, FOXM1 might be a potential prognostic biomarker in ADC.
Acknowledgments
The authors thank Kristofer Wood for revising the manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1103
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1103). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Zhan C, Yan L, Wang L, et al. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J Thorac Dis 2015;7:1398-405. [PubMed]
- Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res 2012;18:1167-76. [Crossref] [PubMed]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 2000;14:142-6. [PubMed]
- Costa RH, Kalinichenko VV, Holterman AX, et al. Transcription factors in liver development, differentiation, and regeneration. Hepatology 2003;38:1331-47. [Crossref] [PubMed]
- Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem 2007;388:1257-74. [Crossref] [PubMed]
- Bhat UG, Halasi M, Gartel AL. Thiazole Antibiotics Target Fox M1 and induce apoptosis in human cancercells. PLoS One 2009;4:e5592. [Crossref] [PubMed]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007;7:847-59. [Crossref] [PubMed]
- Wang IC, Chen YJ, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 2005;25:10875-94. [Crossref] [PubMed]
- Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta 2012;1819:28-37. [Crossref] [PubMed]
- Bella L, Zona S, Nestal de Moraes G, et al. FOXM1: A key oncofoetal transcription factor in health and disease. Semin Cancer Biol 2014;29:32-9. [Crossref] [PubMed]
- Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep 2014;31:2660-8. [Crossref] [PubMed]
- Fei BY, He X, Ma J, Zhang M, Chai R. FoxM1 is associated with metastasis in colorectal cancer through induction of the epithelial-mesenchymal transition. Oncol Lett 2017;14:6553-61. [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang D. W., Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Hou S, Zhou S, Qin Z, et al. Evidence, Mechanism, and Clinical Relevance of the Transdifferentiation from Lung Adenocarcinoma to Squamous Cell Carcinoma. Am J Pathol 2017;187:954-62. [Crossref] [PubMed]
- Han F, Liu W, Xiao H, et al. High expression of SOX30 is associated with favorable survival in human lung adenocarcinoma. Sci Rep 2015;5:13630. [Crossref] [PubMed]
- Han F, Zhang MQ, Liu WB, et al. SOX30 specially prevents Wnt-signaling to suppress metastasis and improve prognosis of lung adenocarcinoma patients. Respir Res 2018;19:241. [Crossref] [PubMed]
- Ribeiro IP, Caramelo F, Esteves L, et al. Genomic and epigenetic signatures associated with survival rate in oral squamous cell carcinoma patients. J Cancer 2018;9:1885-95. [Crossref] [PubMed]
- Elfandy H, Armenia J, Pederzoli F, et al. Genetic and Epigenetic Determinants of Aggressiveness in Cribriform Carcinoma of the Prostate. Mol Cancer Res 2019;17:446-56. [Crossref] [PubMed]
- Ke Y, Zhao W, Xiong J, et al. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem Res Int 2013;2013:506731. [Crossref] [PubMed]
- Ma N, Zhang W, Qiao C, et al. The Tumor Suppressive Role of MiRNA-509-5p by Targeting FOXM1 in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2016;38:1435-46. [Crossref] [PubMed]
- Zeng J, Wang L, Li Q, et al. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27kip1. Journal of Pathology 2009;218:419-27. [Crossref] [PubMed]
- Ma T, Van Tine BA, Wei Y, et al. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 2000;14:2298-313. [Crossref] [PubMed]
- Fukuse T, Hirata T, Naiki H, et al. Prognostic significance of cyclin E overexpression in resected non-small cell lung cancer. Cancer Res 2000;60:242-4. [PubMed]
- Hayashi H, Ogawa N, Ishiwa N, et al. High cyclin E and low p27/Kip1 expressions are potentially poor prognostic factors in lung adenocarcinoma patients. Lung Cancer 2001;34:59-65. [Crossref] [PubMed]
- Wang IC, Ustiyan V, Zhang Y, et al. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 2014;33:5391-6. [Crossref] [PubMed]
- Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001;410:1111-6. [Crossref] [PubMed]
- Zhang J, Niu Y, Huang C. Role of FoxM1 in the Progression and Epithelial to Mesenchymal Transition of Gastrointestinal Cancer. Recent Pat Anticancer Drug Discov 2017;12:247-59. [Crossref] [PubMed]


