
Collagen family genes and related genes might be associated with prognosis of patients with gastric cancer: an integrated bioinformatics analysis and experimental validation
Introduction
Data from the GLOBOCAN database indicates that, globally, there are more than 1,000,000 new cases of gastric cancer (GC) each year, causing an estimated 783,000 deaths in 2018, making it the fifth most frequently diagnosed cancer and the third leading cause of cancer deaths (1). While new treatment strategies and drug developments have made significant progress, due to the low early detection rate of GC, the survival rate of GC patients remains low (2,3). In addition to the existing primary treatments, targeted therapy is expected to be an essential supplementary treatment for advanced GCs (4). Therefore, it is necessary to explore new molecular targets as well as new, highly sensitive and specific biomarkers to elucidate the molecular mechanisms of GC and improve the prognosis of patients with GC.
Recently, bioinformatics analyses (5) have become increasingly popular for analyzing gene expression changes of the in the progression and development of diseases. For example, the online GEO database (http://www.ncbi.nlm.nih.gov/geo) is a public functional genomics tool that can be utilized to analyze experimental gene expression data uploaded by researchers to identify differentially expressed genes (DEGs) of import to disease. The DAVID online database (http://david.ncifcrf.gov) holds information related to proteins and genes, and can be used to mine data for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of these genes. Similarly, differences in gene expression between tumor and normal tissues can be obtained from the TCGA database. STRING (http://string-db.org) is an online database for use in analyzing PPI networks. These online databases assist in experimental data integration and identification of important genes. In the present study, using GO enrichment analysis, we found several DEGs in GC patients, including collagens (COLs), alcohol dehydrogenases (ADHs), N-acetyl galactosyltransferases (GALNTs). Combining the KEGG, GO, and PPI network analysis results, we selected COLs for more in-depth analysis.
From the HUGO Gene Nomenclature Committee database (https://www.genenames.org/), we know that collagen-encoded proteins contain one or more collagen-like domains. Found in vertebrates, this fibrin is a significant component of skin, bones, tendons, cartilage, blood vessels and teeth. Moreover, it is a substantial component of the tumor microenvironment and is involved in cancer fibrosis (6,7). Cancer cells can regulate collagen biosynthesis through mutant genes (8), transcription factors (9,10), signaling pathways and receptors (11,12). Furthermore, collagen can affect tumor cell behavior through tyrosine kinase receptors, integrins, domain receptors, discoidin and some signaling pathways. In GC, collagen type IV α3 chain (COL4A3) has been identified as a potential prognostic factor (13), but few articles have discussed the relationship between collagen genes and GC (14). Therefore, we performed an in-depth study of the COL gene family’s role in GC in order to expose progression mechanisms and to identify prognostic and progression markers.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-1726).
Methods
Microarray data and Identification of DEGs.
We downloaded four gene expression series (GSE79973, GSE26899, GSE54129 and GSE29272) from the GEO database and screened the DEGs of each series between GC and normal samples by GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r). Genes with more than one probe set or probe sets without corresponding gene symbols were removed or averaged, respectively. Adjusted P value <0.01 and |log2Fold Change| >1 were considered statistically significant. Venn diagram of the differentially upregulated and downregulated genes were created (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
KEGG and GO enrichment analyses of DEGs
We used the DAVID (http://david.ncifcrf.gov) online database (version 6.8) to analyze the function of identified DEGs and P<0.05 was considered statistically significant.
PPI network construction and module analysis
In the present study, we used STRING (http://string-db.org) (version 11.0) to construct the PPI network of the DEGs, where a combined score >0.9 was considered statistically significant. We utilized Cytoscape (version 3.7.2) to analyze the molecular interaction networks and MCODE, a Cytoscape app for finding densely connected regions in a given network, was used to identify the most significant modules in the PPI networks. The criteria for selection were as follows: node score cut-off =0.1, degree cut-off =2, k-score =2 and Max depth =100. The genes in the module were analyzed by GO and KEGG using DAVID.
COLs Gene Expression between normal and tumor samples
We utilized Oncomine (https://www.oncomine.org/) to investigate the mRNA levels of COLs in normal and tumor tissues. We retrieved twelve members of COL family genes from the Oncomine database. In our study, the P values of comparison were generated from the student’s t-test. The fold change and cut-off P value were defined as 2 and 0.01, respectively. The expression of COL genes in normal and gastric tumor tissues was also studied using the TCGA-STAD database (http://ualcan.path.uab.edu/index.html).
COL gene methylation between normal versus tumor tissues
We compared the DNA methylation of COL genes between normal and GC tissues using the Human Pan-cancer Methylation database, MethHC (http://methhc.mbc.nctu.edu.tw/). The correlation between COL mRNA expression and the methylation in GC patients was analyzed. In our study, the average value was used as a method for evaluating methylation levels and promoter regions selected for analysis.
Prognostic values of COL members in GC patients
The Kaplan-Meier plotter online database (http://kmplot.com) was used to analyze the relationship between COL expression and the overall survival (OS), first progression (FP), and post-progression survival (PPS) in GC patients. The median COL expression was used as the cut-off. Log-rank P value and hazard ratios, with 95% CI, were calculated.
Cell culture, RNA extraction and real-time quantitative PCR
Human GC cells (AGS, MKN45, HGC27, SGC7901) and human gastric mucosal epithelial cells (GES-1) brought from ATCC were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were maintained at 37 °C in a 5% CO2 atmosphere. Total RNA was extracted from cell samples using an Animal Total RNA Isolation Kit (Foregene, China). After quality control, total RNA was reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). A SYBR Green ITM premix Ex TaqTM II reagent kit (Takara Biomedical Technology, Guangzhou, China) was employed to amplify and quantify the cDNA templates. All PCR reaction systems and conditions were conducted according to the manufacturer's instructions. Primers for COL3A1 were 5'-GAAAGAGGATCTGAGGGCTCC-3' (forward) and 5'-AAACCGCCAGCTTTTTCACC-3' (reverse) and those for COL5A1 were 5'-CTGACAAGAAGTCCGAAGGGG-3' (forward) and 5'-CGTCCACATAGGAGAGCAGTTT-3' (reverse). Primers for β-actin were 5'-AACTGGGACGACATGGAGAAAA-3' (forward) and 5'-GGATAGCACAGCCTGGATAGCA-3' (reverse). The 2–∆∆Ct method was used to calculate expression levels of target genes.
Statistical analysis
Normally distributed data were expressed as mean ± standard deviation (x ± SD). To examine statistical differences between mRNA expression levels and DNA methylation levels of normal and tumor tissues in GC patients, a two-tailed unpaired Student’s t-test was used, P<0.05 was considered to indicate a statistically significant difference. The RT-qPCR analysis was made by GraphPad Prism 7 software and the t-test was used.
Results
Identification of DEGs and GO enrichment and KEGG analyses
After integrating microarray results according to our standards, we identified several DEGs (3160 in GSE54129, 1581 in GSE79973, 428 in GSE26899 and 445 in GSE29272). The overlap among the four gene expression series contained 407 genes, as shown in the Venn diagram (Figure 1), consisting of 275 downregulated genes (Figure 1A) and 132 upregulated genes (Figure 1B). Among the 407 overlapping genes, we used the DAVID online analysis tool to upload all genes that were upregulated and downregulated, thereby determining statistically rich GO terms and KEGG pathways. GO analysis results showed that upregulated DEGs were involved mainly in extracellular matrix (ECM), organization in biological processes (BP), the ECM in cell component (CC), and ECM structural constituent in molecular function (MF). Moreover, downregulated DEGs were involved mainly with ethanol oxidation in BP and ADH activity in MF (Table 1). The significantly enriched pathways of the DEGs analyzed by the KEGG database are shown in Table 2. Upregulated genes were enriched mainly in the ECM-receptor interaction, focal adhesion, protein digestion and absorption, amoebiasis and PI3K-Akt signaling pathway. Downregulated genes were enriched mainly in chemical carcinogenesis, retinol metabolism, glycolysis/gluconeogenesis, metabolism of xenobiotics by cytochrome P450 and drug metabolism-cytochrome P450.
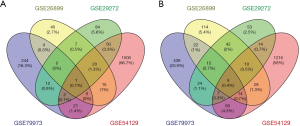
Table 1
| Ontology | ID | Description | P value | Amount | Main gene |
|---|---|---|---|---|---|
| The enriched GO terms of upregulated genes | |||||
| BP | GO:0030198 | Extracellular matrix organization | 7.39E-20 | 23 | COL18A1, COL4A2, COL4A1, OLFML2B, COL3A1 |
| BP | GO:0030574 | Collagen catabolic process | 4.86E-14 | 13 | COL18A1, CTSL, COL4A2, COL4A1, COL3A1 |
| BP | GO:0030199 | Collagen fibril organization | 1.51E-08 | 8 | COL3A1, COL1A2, FOXC1, COL1A1, GREM1 |
| BP | GO:0071230 | Cellular response to amino acid stimulus | 0.00043 | 5 | COL4A1, COL3A1, COL1A2, COL1A1, COL5A2 |
| BP | GO:0030168 | Platelet activation | 0.0018 | 6 | PDPN, COL3A1, COL1A2, FCER1G, COL1A1 |
| BP | GO:0001568 | Blood vessel development | 0.0029 | 4 | SPHK1, COL1A2, COL1A1, COL5A1 |
| CC | GO:0031012 | Extracellular matrix | 3.53E-20 | 26 | ASPN, IGFBP7, COL3A1, SERPINE2, APOE |
| CC | GO:0005581 | Collagen trimer | 4.41E-11 | 12 | COL18A1, CTHRC1, C1QB, COL3A1, COL6A3 |
| CC | GO:0005788 | Endoplasmic reticulum lumen | 1.083E-08 | 13 | COL18A1, COL4A2, COL4A1, COL3A1, COL6A3 |
| MF | GO:0005201 | Extracellular matrix structural constituent | 1.035E-09 | 10 | COL4A2, BGN, COL4A1, COL3A1, COL1A2 |
| MF | GO:0048407 | Platelet-derived growth factor binding | 7.71E-07 | 5 | COL4A1, COL3A1, COL1A2, COL1A1, COL5A1 |
| MF | GO:0046332 | SMAD binding | 0.0035 | 4 | COL3A1, COL1A2, COL5A2, FLNA |
| MF | GO:0004252 | Serine-type endopeptidase activity | 0.0023 | 8 | CTSL, C1QB, BMP1, C3, FAP |
| The enriched GO terms of downregulated genes | |||||
| BP | GO:0006069 | Ethanol oxidation | 0.00036 | 4 | ADH1C, ADH1B, ADH1A, ADH7 |
| BP | GO:0016266 | Glycan processing | 0.000087 | 7 | MUC1, GALNT10, GALNT6, GALNT5, GCNT1 |
| BP | GO:0071985 | Multivesicular body sorting pathway | 0.0089 | 3 | SYTL4, RAB27B, RAB27A |
| MF | GO:0004024 | Alcohol dehydrogenase activity, zinc-dependent | 0.000036 | 4 | ADH1C, ADH1B, ADH1A, ADH7 |
| MF | GO:0016491 | Oxidoreductase activity | 0.000041 | 12 | FAR1, ERO1B, CYP2C9, EGLN3, ADH1C |
| MF | GO:0004745 | Retinol dehydrogenase activity | 0.0013 | 4 | BMP2, ADH1C, ADH1A, ADH7 |
| MF | GO:0004022 | Alcohol dehydrogenase (NAD) activity | 0.0031 | 3 | ADH1C, ADH1A, ADH7 |
| MF | GO:0004653 | Polypeptide N-acetylgalactosaminyltransferase activity | 0.0018 | 4 | GALNT10, GALNT6, GALNT5, GALNT12 |
GO, gene ontology; BP, biological process; CC, cell component; MF, molecular function.
Table 2
| Ontology | ID | Description | P value | Counts | Gene |
|---|---|---|---|---|---|
| The enriched KEGG pathway of upregulated genes | |||||
| KEGGPATHWAY | hsa04512 | ECM-receptor interaction | 8.11E-13 | 14 | COL4A2, COL4A1, COL3A1, COL5A2, COL5A1 |
| KEGGPATHWAY | hsa04510 | Focal adhesion | 4.65E-10 | 16 | COL4A2, COL4A1, COL3A1, COL5A2, FLNA |
| KEGGPATHWAY | hsa04974 | Protein digestion and absorption | 1.08E-07 | 10 | COL18A1, COL4A2, COL4A1, COL3A1, COL6A3 |
| KEGGPATHWAY | hsa05146 | Amoebiasis | 5.42E-07 | 10 | COL4A2, COL4A1, COL3A1, COL1A2, CXCL8 |
| KEGGPATHWAY | hsa04151 | PI3K-Akt signaling pathway | 1.55E-05 | 14 | COL4A2, COL4A1, COL3A1, COL5A2, COL5A1 |
| KEGGPATHWAY | hsa04611 | Platelet activation | 1.35E-03 | 7 | COL3A1, COL1A2, FCER1G, FCGR2A, COL1A1 |
| KEGGPATHWAY | hsa05133 | Pertussis | 6.81E-05 | 7 | C1QB, ITGA5, C3, CXCL8, SERPING1 |
| KEGGPATHWAY | hsa04610 | Complement and coagulation cascades | 4.55E-04 | 6 | C1QB, C3, SERPINE1, SERPING1, C2 |
| KEGGPATHWAY | hsa04610 | Complement and coagulation cascades | 4.55E-04 | 6 | C1QB, C3, SERPINE1, SERPING1, C2 |
| The enriched KEGG pathway of downregulated genes | |||||
| KEGGPATHWAY | hsa05204 | Chemical carcinogenesis | 1.36E-05 | 9 | CYP2C9, CYP2C18, SULT1A1, ADH1C, ADH1B |
| KEGGPATHWAY | hsa00830 | Retinol metabolism | 2.68E-05 | 8 | CYP2C9, CYP2C18, ADH1C, ADH1B, ADH1A |
| KEGGPATHWAY | hsa00010 | Glycolysis / Gluconeogenesis | 3.63E-05 | 8 | LDHB, ALDOB, ADH1C, ADH1B, ADH1A |
| KEGGPATHWAY | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 6.94E-05 | 8 | CYP2C9, ADH1C, ADH1B, ADH1A, ADH7 |
| KEGGPATHWAY | hsa00982 | Drug metabolism - cytochrome P450 | 3.44E-04 | 7 | CYP2C9, ADH1C, ADH1B, ADH1A, ADH7 |
| KEGGPATHWAY | hsa00350 | Tyrosine metabolism | 0.001338831 | 5 | ADH1C, ADH1B, ADH1A, ADH7, ALDH3A1 |
| KEGGPATHWAY | hsa00512 | Mucin type O-Glycan biosynthesis | 8.40E-04 | 5 | GALNT10, GALNT6, GALNT5, GCNT1, GALNT12 |
hsa, Homo sapiens.
DEG PPI network analyses
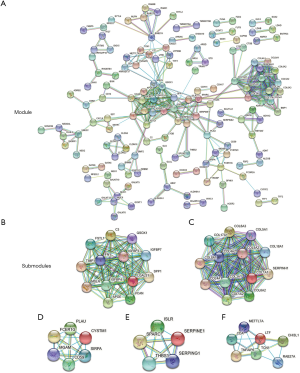
PPI networks involving 150 DEGs (consisting of 75 downregulated genes and 75 upregulated genes) were constructed (Figure 2A), excluding the DEGs which could not constitute a part of a network. With the cut-off criterion set as degrees ≥12, there were 26 genes selected as hub genes, including Quiescin sulfhydl oxidase-1 (QSOX1), Fibronectin-1 (FN1), Tissue inhibitor of metalloproteinases-1 (TIMP1), C3 complement, Collagen 18A1 (COL18A1), Mesothelin (MSLN) and Collagen 1A1 (COL1A1). Cytoscape was used to obtain the 5 most significant submodules (Figure 2B,C,D,E,F). In these submodules, we found several members of the insulin-like growth factor binding protein (IGFBP) gene family (first submodule), collagen (COL) gene family members (second submodule), and serpin peptidase inhibitors (SERPIN) gene family members (fourth submodule). These results suggested that IGFBP, COL and SERPIN family members play an essential role in the development of GC. Functional enrichment results of the second submodule, which involved collagen gene family members, revealed that the development of GC was associated with ECM organization in a biological process, similar to the GO analysis, platelet-derived growth factor binding in MF, and collagen trimer in the cellular component. Other submodules are detailed in Table S1.
Up-regulation of COLs in GC patients
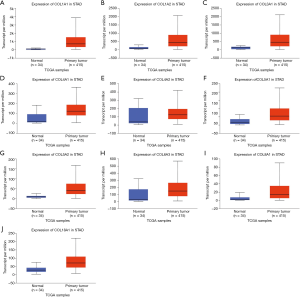
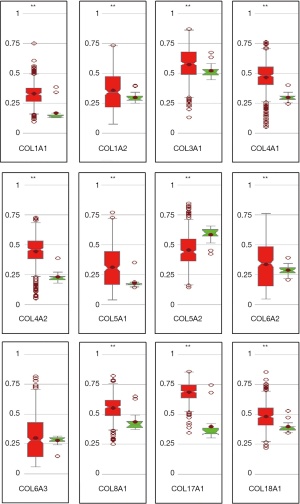
In the GO and KEGG enrichment analysis, several members of the collagen gene family frequently were enriched and, in the PPI network analysis, several COL genes were involved in the second significant submodule. Therefore, COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A1, COL5A2, COL6A2, COL6A3, COL8A1, COL17A1 and COL18A1, which were in the COL gene family and involved in the DEGs, were chosen for more in-depth analysis. To understand better the potential relationship between GC and collagen genes, we used the Oncomine and TCGA-STAD databases to examine the mRNA expression levels of COL genes in normal and gastric tumor tissue. We assessed the expression differences of COLs in 20 cancer samples and their paired normal tissues in the Oncomine database. In these tumor datasets, COL isoforms were significantly upregulated in breast cancer, esophageal cancer, GC, head and neck cancer and pancreatic cancer (Figure 3) compared to matched normal tissues. As the Oncomine and TCGA-STAD databases showed, other COLs were significantly upregulated in tumor tissues (Figures 3,4), except for COL6A2 and COL17A1 (data not shown). The details of COL gene expression in all GC datasets in the Oncomine database are shown in Table S2.

DNA methylation of COL genes in GC patients
In order to explore the role of methylation in the regulation of COL expression in GC patients, the MethHC method was used to analyze the methylation level of the COL genes promoter regions, and the relationship between DNA methylation level and mRNA expression level. Among the COL members, the methylation levels between normal and cancer samples were statistically different (P<0.05, Figure 5), except for COL8A1. Notably, DNA methylation of most COLs (10/11) in GC was higher than in the matched normal tissue, except for COL5A2, which was lower than the normal tissue (Figure 5). The relationship between DNA methylation and mRNA expression of COL members in GC are listed in Table S3, although the R values did not prove the relationship between mRNA level and DNA methylation.
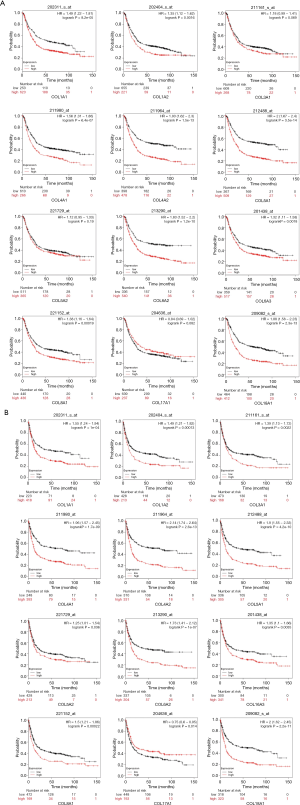
Prognostic characteristics of COLs in GC patients
Prognostic characteristics of GC patients, including OS, first progression (FP), and post progression survival (PPS), were surveyed in the Kaplan-Meier plotter database. Among these COLs available in the Kaplan-Meier database, most genes showed a positive relationship between high expression and significantly worse OS in GC patients (Figure 6A), except COL3A1, COL5A2 and COL17A1. The data showed FP reduction with low COL17A1 (Figure 6B) and high levels of the other collagen genes. The significant, inverse relationship was shown between PPS and collagen genes, except for COL5A2 and COL17A1 (Figure 6C). High COL1A1, COL1A2, COL4A1, COL4A2, COL5A1, COL6A2, COL6A3, COL8A1 and COL18A1 mRNA expression levels led to reduced OS, FP and PPS in GC patients. Furthermore, increased COL17A1 mRNA levels significantly correlated only with increased FP, but was not correlated with OS or PPS. In Lauren’s classification, GC is divided into three categories: diffuse, intestinal and mixed. Therefore, the Kaplan-Meier plotter online tool can be used to determine the prognostic value of COL gene isoforms in different GC subtypes. The data showed that high expression levels of COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A1, COL6A2, COL6A3, and COL18A1 led to reduced OS, FP and PPS in intestinal and diffuse-type GC patients. Additionally, in the mixed-type GC patients, most of the COLs were with no-significance because the number of the cases were too small for statistical analysis (Table S4). The different transcript levels of COL17A1 had no effect on the three subtypes, except the OS in intestinal type, which corresponded to the result where the COL17A1 mRNA expression level showed no difference between the normal and tumor tissues. The complex relationship of these GC subtype survival time (OS, FP, PPS) with the COLmRNA expression was shown in the supplementary materials (Figures S1-S3).
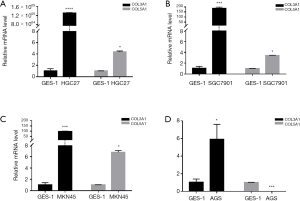
mRNA expression of COL3A1 and COL5A1 in different GC cells
Except for COL6A2 and COL17A1, COLs were highly expressed according to the TCGA-STAD databases. We chose the COL3A1 and COL5A1 genes, which lacked experimental verification in GC but have already been shown to play roles in other cancers (15,16), for RT-qPCR experiments to validate our analysis results. As shown in Figure 7, the COL3A1 level was 1.310×106 folds higher in HGC27, 185 folds higher in SGC7901, 96 folds higher in MKN45 and six folds higher in AGS human GC cell lines, compared with GES-1 normal human gastric mucosal epithelial cell line, and these results were consistent with the analyses from the TCGA-STAD databases. Interestingly, COL5A1 was highly expressed in HGC27, MKN45 and SGC7901, at about 3-7 folds, which were also consistent with the above results. However, in the AGS cell lines, COL5A1 was 400 folds lower than in GES-1. These results require additional in-depth exploration.
Discussion
In recent years, significant efforts have been made in order to understand better the early diagnosis, targeted therapy and prognosis of GC (17,18). However, the OS of patients with GC remains unimproved, particularly in developing countries (19,20). Our study aimed to identify nuclear genes with similar functions that are highly expressed in GC, compared to normal controls, and to reveal their underlying mechanisms. In the present study, we downloaded the gene expression series of GSE79973, GSE26899, GSE54129 and GSE29272 from the GEO database and found 132 upregulated and 275 downregulated overlap DEGs between GC and normal controls. GO term analysis showed that upregulated DEGs were related primarily with ECM. As reported in previous studies, several ECM-related genes had impacts on the development of GC (21-23). Increased deposition of matrix proteins favors tumor progression by interfering with cell polarity, cell-cell adhesion and, ultimately, amplifying growth factor signaling. As the most significant ECM component (24), collagen determines the functional properties of the matrix and changes in the deposition or degradation of collagen can lead to a decline of ECM homeostasis. It has been reported that increased collagen cross-linking and deposition leads to tumor progression via increased integrin signaling (25). PPI network analysis showed that the IGFBP, SERPIN, and COL gene families were enriched in several submodules. Previous studies have shown that IGFBPs play a protective role in the process of GC development (26-28). However, in our meta-analysis, we found that IGFBP3, IGFBP4, and IGFBP7 were upregulated in GC patients, which was opposed to normal tissues. This might be a self-protection mechanism in GC patients, and additional experiments and analyses are required to investigate this unusual situation. Wang et al. (29), Ju et al. (30), and Yang et al. (31) found that SERPINs can be used as a novel prognostic factor in GC. Additionally, Tian et al. found that SERPINH1 was overexpressed in GC patients and took part in the regulation of EMT (32), which supported the results of our analysis.
Among the identified DEGs, 12 collagen genes were found. Most of these collagen genes with high mRNA and DNA methylation levels ExceptingCOL6A2, COL8A1, COL17A1 and COL5A2, these collagen genes were found to have high mRNA and DNA methylation levels. DNA methylation causes gene silencing. Our results showed, however, high DNA methylation in the promoter region (except for COL5A2), similar to the mRNA levels, in the GC cells. The results showed that methylation in the promotor region did not influence mRNA expression levels COL genes and suggested that methylation may exist in another region or some other mechanism may have affected mRNA levels. Kaplan-Meier analysis revealed that most of the COLs showed a positive relationship between high expression and significantly worse prognoses in GC patients, which supported the idea that COLs could be prognostic markers in GC patients. Previously, only a few isoforms of COLs involved in GC were reported. Previous studies have demonstrated that upregulated expression of COL1A1 (33), COL1A2 and COL6A3 (34) enhanced the invasive properties of GC cells. COL4A3 was confirmed as a prognostic factor in GC (13). The role of other COLs in GC has not been published (14). Accordingly, additional experimental verification is required to confirm our results and evaluate their meaning. It has been shown that COL3A1 and COL5A1 can be a diagnostic marker in breast cancer and plays a role in non-small cell lung cancer (15,16,35). Therefore, we chose these two COLs for RT-qPCR experiments. After our repeated experiments, data showed that COL3A1 was highly expressed in the four cell lines, and that COL5A1 was highly expressed, in except AGS cells. The differing expression levels between GC cell lines suggested to determine the differences between the cell lines. We found that between these four GC cell lines, HGC27 had the highest degree of malignancy, while AGS was the lowest (36-39).These results are consistent with the expression levels of COL3A1 and COL5A1 in each of the cell lines, which provided a basis for COL3A1 and COL5A1 as markers for the progression and prognosis of GC.
Conclusions
Additional experimentation is required in order to determine whether the COL gene family can be utilized as markers of GC progression and prognosis. Our analysis provides a feasible basis for the idea that COLs may be used as progression and prognosis markers of GC.
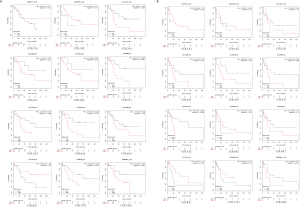
Table S1
| Characteristics | Nodes | Edges | Average node degree | Average local clustering coefficient | PPI enrichment P value | Key genes | Functional enrichment | ||
|---|---|---|---|---|---|---|---|---|---|
| BP | MF | CC | |||||||
| Module | 365 | 371 | 2.03 | 0.36 | <1.0e-16 | QSOX1, FN1, TIMP1, C3, MSLN | Extracellular structure organization | Platelet-derived growth factor binding | Endoplasmic reticulum lumen |
| Submodules | |||||||||
| 1 | 13 | 78 | 12 | 1 | <1.0e-16 | IGFBP7, IGFBP3, QSOX1, VCAN, TIMP1 | Post-translational protein modification | Insulin-like growth factor binding | Endoplasmic reticulum lumen |
| 2 | 13 | 78 | 12 | 1 | <1.0e-16 | COL1A1, COL1A2, SERPINH1, COL17A1, COL4A2 | Extracellular matrix organization | Platelet-derived growth factor binding | Collagen trimer |
| 3 | 6 | 15 | 5 | 1 | <1.0e-16 | MGAM, PLAU, SIRPA, FCER1G, CYSTM1 | Neutrophil degranulation | - | Tertiary granule membrane |
| 4 | 5 | 10 | 4 | 1 | 2.03E-13 | SERPINE1, SERPING1, THBS1, ISLR, SPARC | Platelet degranulation | Extracellular matrix binding | Platelet alpha granule lumen |
| 5 | 7 | 15 | 4.29 | 0.886 | <1.0e-16 | METTL7A, CHI3L1, LTF, TCN1, TNFAIP6 | Neutrophil degranulation | Carbohydrate derivative binding | Tertiary granule lumen |
Nodes, the gene numbers in the modules. Edges, the interaction numbers in the modules. PPI enrichment P value indicate that the nodes are not random and that the observed number of edges is significant.
Table S2
| Collagen family | Types of Gastric Cancer vs. normal | Fold change | t-test | P value | Reporter |
|---|---|---|---|---|---|
| COL1A1 | Gastric Cancer vs. Normal | 3.201 | 8.724 | 1.81E-15 | Cui Gastric Statistics;3762198 |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 5.483 | 12.628 | 3.47E-21 | Chen Gastric Statistics; IMAGE:153646 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 9.047 | 8.649 | 1.65E-07 | Chen Gastric Statistics IMAGE:418193 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 11.917 | 7.514 | 1.90E-05 | Chen Gastric Statistics; IMAGE:153647 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 5.607 | 7.715 | 4.58E-10 | Cho Gastric Statistics ILMN_1701308 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 4.081 | 5.196 | 4.71E-06 | Cho Gastric Statistics ILMN_1701308 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.652 | 3.295 | 0.002 | Cho Gastric Statistics ILMN_1701308 | |
| Gastric Cancer vs. Normal | 5.808 | 6.795 | 2.99E-06 | Wang Gastric Statistics;202310_s_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 6.017 | 8.766 | 5.20E-11 | DErrico Gastric Statistics 202311_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 5.538 | 5.334 | 8.77E-04 | DErrico Gastric Statistics 202311_s_at | |
| Gastric Mixed Adenocarcinoma vs. Normal | 5.471 | 8.023 | 6.74E-04 | DErrico Gastric Statistics 202310_s_at | |
| COL1A2 | Gastric Cancer vs. Normal | 7.491 | 7.308 | 2.81E-07 | Wang Gastric Statistics; 202404_s_AT |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 4.548 | 15.552 | 6.07E-25 | Chen Gastric Statistics;IMAGE839991 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 5.193 | 11.394 | 2.23E-10 | Chen Gastric Statistics;IMAGE839991 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 6.984 | 8.952 | 4.51E-05 | Chen Gastric Statistics;IMAGE839991 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 5.876 | 9.253 | 1.89E-12 | Cho Gastric Statistics;ILMN_2104356 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 4.062 | 5.226 | 9.69E-06 | Cho Gastric Statistics;ILMN_2104356 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 3.404 | 4.337 | 4.87E-04 | Cho Gastric Statistics;ILMN_2104356 | |
| Gastric Cancer vs. Normal | 2.277 | 7.245 | 9.49E-12 | Cui Gastric Statistics;3013054 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 7.433 | 10.405 | 5.42E-14 | DErrico Gastric Statistics;202404_s_at | |
| Gastric Mixed Adenocarcinoma vs. Normal | 3.453 | 9.816 | 2.37E-08 | DErrico Gastric Statistics;202403_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 6.424 | 5.785 | 4.63E-04 | DErrico Gastric Statistics;202404_s_at | |
| COL3A1 | Gastric Cancer vs. Normal | 2.333 | 7.397 | 4.15E-12 | Cui Gastric Statistics;2519577 |
| Diffuse Gastric Adenocarcinoma vs. Normal | 4.458 | 10.994 | 2.57E-11 | Chen Gastric Statistics IMAGE:122159(1) | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.466 | 12.075 | 5.06E-19 | Chen Gastric Statistics IMAGE:122159(2) | |
| Gastric Mixed Adenocarcinoma vs. Normal | 5.675 | 8.902 | 2.65E-06 | Chen Gastric Statistics IMAGE:122159(2) | |
| Gastric Cancer vs. Normal | 2.766 | 6.322 | 2.41E-06 | Wang Gastric Statistics;215076_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.656 | 5.201 | 2.04E-06 | Cho Gastric Statistics;ILMN_1773079 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.225 | 3.13 | 0.002 | Cho Gastric Statistics;ILMN_1773079 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.864 | 7.241 | 2.31E-05 | DErrico Gastric Statistics;215076_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.7 | 4.516 | 9.72E-04 | DErrico Gastric Statistics;201852_s_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.425 | 5.784 | 2.29E-07 | DErrico Gastric Statistics;215076_s_at | |
| COL4A1 | Diffuse Gastric Adenocarcinoma vs. Normal | 5.045 | 14.254 | 4.54E-13 | Chen Gastric Statistics;IMAGE:145292 |
| Gastric Mixed Adenocarcinoma vs. Normal | 6.23 | 10.438 | 6.43E-07 | Chen Gastric Statistics;IMAGE:145292 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 4.104 | 15.779 | 6.04E-18 | Chen Gastric Statistics;IMAGE:145292 | |
| Gastric Cancer vs. Normal | 2.276 | 5.853 | 5.67E-06 | Wang Gastric Statistics;211980_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.207 | 10.716 | 7.08E-14 | DErrico Gastric Statistics;211980_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 3.705 | 3.88 | 0.002 | DErrico Gastric Statistics;211981_at | |
| Gastric Mixed Adenocarcinoma vs. Normal | 7.956 | 6.055 | 0.001 | DErrico Gastric Statistics;211981_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.884 | 6.164 | 3.10E-07 | Cho Gastric Statistics;ILMN_1653028 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.25 | 3.838 | 2.35E-04 | Cho Gastric Statistics;ILMN_1653028 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.676 | 3.972 | 4.37E-04 | Cho Gastric Statistics;ILMN_1653028 | |
| COL4A2 | Diffuse Gastric Adenocarcinoma vs. Normal | 3.14 | 10.501 | 1.63E-09 | Chen Gastric Statistics;IMAGE:769959 |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.133 | 10.669 | 2.38E-17 | Chen Gastric Statistics;IMAGE:769959 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 3.82 | 9.131 | 4.23E-06 | Chen Gastric Statistics;IMAGE:769959 | |
| Gastric Cancer vs. Normal | 2.483 | 5.93 | 1.85E-06 | Wang Gastric Statistics;211964_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.632 | 9.368 | 3.54E-12 | DErrico Gastric Statistics;211964_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 3.047 | 3.886 | 0.002 | DErrico Gastric Statistics;211966_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.425 | 5.644 | 4.41E-07 | Cho Gastric Statistics ILMN_1724994 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.515 | 4.902 | 1.57E-05 | Cho Gastric Statistics ILMN_1724994 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.618 | 3.618 | 0.002 | Cho Gastric Statistics ILMN_1724994 | |
| COL5A1 | Gastric Cancer vs. Normal | 3.946 | 7.332 | 5.77E-07 | Wang Gastric Statistics;212488_at |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.981 | 6.907 | 1.45E-08 | DErrico Gastric Statistics;203325_s_at | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.714 | 6.924 | 7.43E-04 | DErrico Gastric Statistics;212488_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.625 | 3.883 | 0.005 | DErrico Gastric Statistics;212488_at | |
| COL5A2 | Gastric Cancer vs. Normal | 2.294 | 8.708 | 2.52E-15 | Cui Gastric Statistics;2591643 |
| Gastric Cancer vs. Normal | 3.287 | 5.94 | 2.89E-06 | Wang Gastric Statistics;221730_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.534 | 12.611 | 2.05E-17 | Chen Gastric Statistics;IMAGE429203 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 3.589 | 7.761 | 4.05E-07 | Chen Gastric Statistics;IMAGE429203 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 4.988 | 7.028 | 4.00E-05 | Chen Gastric Statistics;IMAGE429203 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 3.393 | 5.78 | 3.98E-07 | Cho Gastric Statistics;ILMN_1729117 | |
| Gastric Adenocarcinoma vs. Normal | 3.03 | 3.59 | 0.007 | Cho Gastric Statistics;ILMN_1729117 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.54 | 3.819 | 2.53E-04 | Cho Gastric Statistics;ILMN_1729117 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.166 | 3.229 | 0.002 | Cho Gastric Statistics;ILMN_1729117 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.77 | 7.502 | 8.64E-09 | DErrico Gastric Statistics;221730_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.885 | 4.174 | 0.003 | DErrico Gastric Statistics;221730_at | |
| COL6A2 | Gastric Cancer vs. Normal | 2.819 | 6.496 | 5.69E-07 | Wang Gastric Statistics;209156_s_at |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.668 | 4.98 | 7.94E-04 | DErrico Gastric Statistics;209156_s_at | |
| COL6A3 | Gastric Cancer vs. Normal | 5.087 | 7.295 | 6.06E-08 | Wang Gastric Statistics;201438_at |
| Gastric Mixed Adenocarcinoma vs. Normal | 5.37 | 16.537 | 1.25E-13 | DErrico Gastric Statistics;201438_at | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.92 | 8.91 | 9.71E-12 | DErrico Gastric Statistics;201438_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 4.619 | 6.311 | 2.79E-04 | DErrico Gastric Statistics;201438_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 3.409 | 8.89 | 9.13E-12 | Cho Gastric Statistics;ILMN_1706643 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.819 | 5.326 | 8.78E-06 | Cho Gastric Statistics;ILMN_1706643 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.583 | 3.174 | 0.003 | Cho Gastric Statistics;ILMN_2307861 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 4.552 | 10.63 | 1.09E-07 | Chen Gastric Statistics;IMAGE:138991 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 3.131 | 11.751 | 4.40E-19 | Chen Gastric Statistics;IMAGE:138991 | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 4.647 | 9.783 | 5.85E-09 | Chen Gastric Statistics;IMAGE:138991 | |
| Gastric Cancer vs. Normal | 2.021 | 5.82 | 1.60E-08 | Cui Gastric Statistics;2605321 | |
| COL8A1 | Diffuse Gastric Adenocarcinoma vs. Normal | 5.2 | 9.378 | 3.63E-12 | Cho Gastric Statistics;ILMN_2402392 |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 4.754 | 7.45 | 4.92E-08 | Cho Gastric Statistics;ILMN_1685433 | |
| Gastric Adenocarcinoma vs. Normal | 6.156 | 5.203 | 0.005 | Cho Gastric Statistics;ILMN_1685433 | |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.759 | 4.424 | 4.73E-04 | Cho Gastric Statistics;ILMN_2402392 | |
| Gastric Cancer vs. Normal | 2.735 | 6.252 | 1.81E-09 | Cui Gastric Statistics;2633390 | |
| Gastric Cancer vs. Normal | 5.094 | 4.553 | 7.70E-05 | Wang Gastric Statistics;214589_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.055 | 3.461 | 0.001 | DErrico Gastric Statistics;221152_at | |
| COL17A1 | Gastric Mixed Adenocarcinoma vs. Normal | -3.494 | -6.925 | 9.86E-08 | Chen Gastric Statistics;IMAGE:252259 |
| Diffuse Gastric Adenocarcinoma vs. Normal | -2.167 | -4.414 | 7.09E-05 | Chen Gastric Statistics;IMAGE:501981 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | -2.447 | -5.491 | 1.16E-06 | Chen Gastric Statistics;IMAGE:252259 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | -2.469 | -2.936 | 0.002 | DErrico Gastric Statistics;204636_at | |
| COL18A1 | Gastric Cancer vs. Normal | 3.074 | 7.036 | 1.14E-07 | Wang Gastric Statistics;209081_s_at |
| Gastric Mixed Adenocarcinoma vs. Normal | 2.233 | 7.572 | 3.30E-05 | Chen Gastric Statistics;IMAGE:301061 | |
| Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.111 | 8.088 | 6.14E-11 | DErrico Gastric Statistics;209081_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.155 | 4.947 | 0.001 | DErrico Gastric Statistics;209082_s_at | |
| Diffuse Gastric Adenocarcinoma vs. Normal | 2.118 | 4.757 | 1.02E-05 | Cho Gastric Statistics;ILMN_1806733 |
Notes: P value was analyzed using the
Table S3
| Gene name | COL1A1 | COL1A2 | COL3A1 | COL4A1 | COL4A2 | COL5A1 | COL5A2 | COL6A2 | COL6A3 | COL8A1 | COL17A1 | COL18A1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R value | −0.0634 | −0.162 | 0.0552 | −0.114 | −0.24 | −0.157 | 0.0779 | −0.128 | 0.0166 | −0.259 | −0.262 | −0.00749 |
| P value | 0 | 0 | 3.33E-16 | 0 | 0 | 0 | 0 | 0.341 | 0 | 3.33E-16 | 0.259 | 0 |
Table S4
| Collagen family | Lauren classification | OS | PPS | FP | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | HR | 95% CI | P value | Cases | HR | 95% CI | P value | Cases | HR | 95% CI | P value | ||||
| COL1A1 | Intestinal | 320 | 2.08 | 1.43–3.02 | 8.70E-05 | 192 | 2.27 | 1.39–3.71 | 0.0007 | 263 | 1.97 | 1.33–2.92 | 0.00052 | ||
| Diffuse | 241 | 1.52 | 1.05–2.21 | 0.026 | 176 | 1.86 | 1.26–2.74 | 0.0016 | 231 | 1.56 | 1.07–2.29 | 0.0202 | |||
| Mixed | 32 | 0.62 | 0.2–1.97 | 0.417 | 16 | – | – | – | 28 | 0.32 | 0.06–1.74 | 0.1643 | |||
| COL1A2 | Intestinal | 320 | 1.79 | 1.31–2.46 | 0.00026 | 192 | 1.6 | 1.04–2.47 | 0.032 | 263 | 1.98 | 1.38–2.82 | 0.00014 | ||
| Diffuse | 241 | 1.71 | 1.21–2.42 | 0.002 | 176 | 2.13 | 1.44–3.16 | 0.00011 | 231 | 1.64 | 1.16–2.32 | 0.005 | |||
| Mixed | 32 | 0.4 | 0.11–1.47 | 0.154 | 16 | – | – | – | 28 | 0.63 | 0.21–1.86 | 0.3955 | |||
| COL3A1 | Intestinal | 320 | 1.49 | 1.09–2.05 | 0.0123 | 192 | 2.02 | 1.34–3.06 | 0.0006 | 263 | 1.93 | 1.35–2.77 | 0.00028 | ||
| Diffuse | 241 | 1.42 | 1.01–1.99 | 0.045 | 176 | 2.02 | 1.38–2.96 | 0.00022 | 231 | 1.47 | 1.04-2.08 | 0.029 | |||
| Mixed | 32 | 2.82 | 1–7.99 | 0.042 | 16 | – | – | – | 28 | 1.53 | 0.57–4.15 | 0.3988 | |||
| COL4A1 | Intestinal | 320 | 1.63 | 1.15–2.32 | 0.0056 | 192 | 1.76 | 1.15–2.69 | 0.0082 | 263 | 2 | 1.36–2.95 | 0.00033 | ||
| Diffuse | 241 | 2.08 | 1.4–3.1 | 0.00022 | 176 | 1.99 | 1.26–3.12 | 0.0024 | 231 | 2.17 | 1.5–3.16 | 2.90E-05 | |||
| Mixed | 32 | 1.97 | 0.55–7.09 | 0.2922 | 16 | – | – | – | 28 | 2.21 | 0.79–6.17 | 0.1222 | |||
| COL4A2 | Intestinal | 320 | 2.58 | 1.81–3.67 | 5.60E-08 | 192 | 2.89 | 1.91–4.37 | 1.60E-07 | 263 | 2.38 | 1.64–3.46 | 2.50E-06 | ||
| Diffuse | 241 | 2.45 | 1.72–3.49 | 3.40E-07 | 176 | 2.63 | 1.77–3.89 | 5.60E-07 | 231 | 2.7 | 1.83–3.98 | 2.10E-07 | |||
| Mixed | 32 | 2.2 | 0.49–9.93 | 0.29 | 16 | – | – | – | 28 | 2.66 | 0.6–11.81 | 0.1811 | |||
| COL5A1 | Intestinal | 320 | 2.55 | 1.85–3.5 | 2.50E-09 | 192 | 2.79 | 1.85–4.22 | 3.80E-07 | 263 | 2.25 | 1.58–3.2 | 3.30E-06 | ||
| Diffuse | 241 | 1.86 | 1.31–2.64 | 0.00043 | 176 | 2.46 | 1.67–3.67 | 2.60E-06 | 231 | 1.82 | 1.27–2.58 | 0.00083 | |||
| Mixed | 32 | 3.83 | 1.35–10.87 | 0.0067 | 16 | – | – | – | 28 | 2.33 | 0.86–6.35 | 0.0892 | |||
| COL5A2 | Intestinal | 320 | 1.49 | 1.07–2.08 | 0.0167 | 192 | 1.34 | 0.89–2.02 | 0.1596 | 263 | 1.51 | 1.04–2.18 | 0.029 | ||
| Diffuse | 241 | 1.26 | 0.89–1.79 | 0.1984 | 176 | 1.54 | 1.03–2.32 | 0.036 | 231 | 1.31 | 0.93–1.85 | 0.118 | |||
| Mixed | 32 | 2.45 | 0.84–7.12 | 0.0886 | 16 | – | – | – | 28 | 4.83 | 1.46–15.94 | 0.0048 | |||
| COL6A2 | Intestinal | 320 | 2.78 | 1.98–3.91 | 8.10E-10 | 192 | 3.54 | 2.33–5.39 | 3.50E-10 | 263 | 2.17 | 1.52–3.09 | 1.40E-05 | ||
| Diffuse | 241 | 2.13 | 1.48–3.06 | 3.10E-05 | 176 | 2.51 | 1.7–3.72 | 2.00E-06 | 231 | 2.08 | 1.43–3.01 | 8.00E-05 | |||
| Mixed | 32 | 3.58 | 1.26–10.22 | 0.0111 | 16 | – | – | – | 28 | 0.52 | 0.16–1.65 | 0.2597 | |||
| COL6A3 | Intestinal | 320 | 2.26 | 1.65–3.1 | 1.90E-07 | 192 | 2.51 | 1.67–3.79 | 5.40E-06 | 263 | 1.95 | 1.37–2.79 | 0.0002 | ||
| Diffuse | 241 | 1.43 | 1–2.03 | 0.048 | 176 | 1.92 | 1.29–2.86 | 0.0011 | 231 | 1.56 | 1.06–2.31 | 0.024 | |||
| Mixed | 32 | 1.77 | 0.62–5.03 | 0.279 | 16 | – | – | – | 28 | 1.71 | 0.61–4.73 | 0.3001 | |||
| COL8A1 | Intestinal | 320 | 1.94 | 1.38–2.72 | 8.80E-05 | 192 | 2.19 | 1.43–3.35 | 0.0002 | 263 | 1.57 | 0.99–2.48 | 0.054 | ||
| Diffuse | 241 | 1.31 | 0.93–1.86 | 0.13 | 176 | 2.21 | 1.5–3.25 | 3.90E-05 | 231 | 1.31 | 0.93–1.86 | 0.1209 | |||
| Mixed | 32 | 0.39 | 0.13–1.25 | 0.1012 | 16 | – | – | – | 28 | 0.4 | 0.14–1.1 | 0.0674 | |||
| COL17A1 | Intestinal | 320 | 0.69 | 0.5–0.94 | 0.0181 | 192 | 1.27 | 0.83–1.95 | 0.2725 | 263 | 0.71 | 0.5–1 | 0.051 | ||
| Diffuse | 241 | 0.72 | 0.47–1.09 | 0.1189 | 176 | 1.28 | 0.83–1.98 | 0.26 | 231 | 0.69 | 0.46–1.05 | 0.0815 | |||
| Mixed | 32 | 2.84 | 0.94–8.53 | 0.053 | 16 | – | – | – | 28 | 0.63 | 0.21–1.87 | 0.4016 | |||
| COL18A1 | Intestinal | 320 | 2.76 | 2.01–3.8 | 7.90E-11 | 192 | 3.14 | 2.08–4.76 | 1.20E-08 | 263 | 2.3 | 1.61–3.3 | 2.90E-06 | ||
| Diffuse | 241 | 1.76 | 1.22–2.53 | 0.002 | 176 | 1.97 | 1.34–2.89 | 0.00042 | 231 | 1.75 | 1.21–2.54 | 0.0025 | |||
| Mixed | 32 | 2.59 | 0.81–8.31 | 0.0967 | 16 | – | – | – | 28 | 2/36 | 0.84–6.6 | 0.0935 | |||
Notes: P value was analyzed using the survival analysis test. OS, overall survival; PPS, post-progression survival; FP, first progression; HR, hazard ratio.
Acknowledgments
We thank the contributions of the staff of the Institute of Antibody Engineering, School of Laboratory Medicine and Biotechnology, Southern Medical University in Guangzhou, China.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1726
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1726). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhang H, Wang X, Huang H, et al. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol 2019;47:308-18. [Crossref] [PubMed]
- Wang YN, Xu F, Zhang P, et al. MicroRNA-575 regulates development of gastric cancer by targeting PTEN. Biomed Pharmacother 2019;113:108716. [Crossref] [PubMed]
- Shah MA, Xu RH, Bang YJ, et al. HELOISE: Phase IIIb Randomized Multicenter Study Comparing Standard-of-Care and Higher-Dose Trastuzumab Regimens Combined With Chemotherapy as First-Line Therapy in Patients With Human Epidermal Growth Factor Receptor 2-Positive Metastatic Gastric or Gastroesophageal Junction Adenocarcinoma. J Clin Oncol 2017;35:2558-67. [Crossref] [PubMed]
- Sedlazeck FJ, Lee H, Darby CA, et al. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet 2018;19:329-46. [Crossref] [PubMed]
- Discher DE, Smith L, Cho S, et al. Matrix Mechanosensing: From Scaling Concepts in 'Omics Data to Mechanisms in the Nucleus, Regeneration, and Cancer. Annu Rev Biophys 2017;46:295-315. [Crossref] [PubMed]
- Yamauchi M, Barker TH, Gibbons DL, et al. The fibrotic tumor stroma. J Clin Invest 2018;128:16-25. [Crossref] [PubMed]
- Wörmann SM, Song L, Ai J, et al. Loss of P53 Function Activates JAK2-STAT3 Signaling to Promote Pancreatic Tumor Growth, Stroma Modification, and Gemcitabine Resistance in Mice and Is Associated With Patient Survival. Gastroenterology 2016;151:180-93.e12. [Crossref] [PubMed]
- Yoshida T, Hashimura M, Kuwata T, et al. Transcriptional regulation of the alpha-1 type II collagen gene by nuclear factor B/p65 and Sox9 in the chondrocytic phenotype of uterine carcinosarcomas. Hum Pathol 2013;44:1780-8. [Crossref] [PubMed]
- Nagathihalli NS, Castellanos JA, Shi C, et al. Signal Transducer and Activator of Transcription 3, Mediated Remodeling of the Tumor Microenvironment Results in Enhanced Tumor Drug Delivery in a Mouse Model of Pancreatic Cancer. Gastroenterology 2015;149:1932-43.e9. [Crossref] [PubMed]
- Laklai H, Miroshnikova YA, Pickup MW, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med 2016;22:497-505. [Crossref] [PubMed]
- Miskolczi Z, Smith MP, Rowling EJ, et al. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene 2018;37:3166-82. [Crossref] [PubMed]
- Nie XC, Wang JP, Zhu W, et al. COL4A3 expression correlates with pathogenesis, pathologic behaviors, and prognosis of gastric carcinomas. Hum Pathol 2013;44:77-86. [Crossref] [PubMed]
- Xu S, Xu H, Wang W, et al. The role of collagen in cancer: from bench to bedside. J Transl Med 2019;17:309. [Crossref] [PubMed]
- Wang Y, Resnick MB, Lu S, et al. Collagen type III alpha1 as a useful diagnostic immunohistochemical marker for fibroepithelial lesions of the breast. Hum Pathol 2016;57:176-81. [Crossref] [PubMed]
- Souza P, Rizzardi F, Noleto G, et al. Refractory remodeling of the microenvironment by abnormal type V collagen, apoptosis, and immune response in non-small cell lung cancer. Hum Pathol 2010;41:239-48. [Crossref] [PubMed]
- Badiyan SN, Hallemeier CL, Lin SH, et al. Proton beam therapy for gastrointestinal cancers: past, present, and future. J Gastrointest Oncol 2018;9:962-71. [Crossref] [PubMed]
- Tan AC, Chan DL, Faisal W, et al. New drug developments in metastatic gastric cancer. Therap Adv Gastroenterol 2018;11:1756284818808072. [Crossref] [PubMed]
- Goetze OT, Al-Batran SE, Chevallay M, et al. Multimodal treatment in locally advanced gastric cancer. Updates Surg 2018;70:173-9. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Umeda S, Kanda M, Miwa T, et al. Fraser extracellular matrix complex subunit 1 promotes liver metastasis of gastric cancer. Int J Cancer 2020;146:2865-76. [Crossref] [PubMed]
- Wang H, Chen H, Jiang Z, et al. Integrin subunit alpha V promotes growth, migration, and invasion of gastric cancer cells. Pathol Res Pract 2019;215:152531. [Crossref] [PubMed]
- Wu H, Qiao F, Zhao Y, et al. Downregulation of Long Non-coding RNA FALEC Inhibits Gastric Cancer Cell Migration and Invasion Through Impairing ECM1 Expression by Exerting Its Enhancer-Like Function. Front Genet 2019;10:255. [Crossref] [PubMed]
- Yan Q, Sui W, Xie S, et al. Expression and role of integrin-linked kinase and collagen IV in human renal allografts with interstitial fibrosis and tubular atrophy. Transpl Immunol 2010;23:1-5. [Crossref] [PubMed]
- Walker C, Mojares E, Del RHA. Role of Extracellular Matrix in Development and Cancer Progression. Int J Mol Sci 2018;19:3028. [Crossref] [PubMed]
- Luo C, Sun F, Zhu H, et al. Insulin-like growth factor binding protein-1 (IGFBP-1) upregulated by Helicobacter pylori and is associated with gastric cancer cells migration. Pathol Res Pract 2017;213:1029-36. [Crossref] [PubMed]
- Kim J, Kim WH, Byeon SJ, et al. Epigenetic Downregulation and Growth Inhibition of IGFBP7 in Gastric Cancer. Asian Pac J Cancer Prev 2018;19:667-75. [PubMed]
- Kim ST, Jang HL, Lee J, et al. Clinical Significance of IGFBP-3 Methylation in Patients with Early Stage Gastric Cancer. Transl Oncol 2015;8:288-94. [Crossref] [PubMed]
- Wang K, Wang B, Xing AY, et al. Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells. J Cancer Res Clin Oncol 2015;141:805-12. [Crossref] [PubMed]
- Ju H, Lim B, Kim M, et al. SERPINE1 intron polymorphisms affecting gene expression are associated with diffuse-type gastric cancer susceptibility. Cancer 2010;116:4248-55. [Crossref] [PubMed]
- Yang J, Xiong X, Wang X, et al. Identification of peptide regions of SERPINA1 and ENOSF1 and their protein expression as potential serum biomarkers for gastric cancer. Tumour Biol 2015;36:5109-18. [Crossref] [PubMed]
- Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/beta-catenin signaling pathway. Aging (Albany NY) 2020;12:3574-93. [Crossref] [PubMed]
- Shi Y, Duan Z, Zhang X, et al. Down-regulation of the let-7i facilitates gastric cancer invasion and metastasis by targeting COL1A1. Protein Cell 2019;10:143-8. [Crossref] [PubMed]
- Ao R, Guan L, Wang Y, et al. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J Cell Biochem 2018;119:4420-34. [Crossref] [PubMed]
- Pan J, Mor G, Ju W, et al. Viral Infection-Induced Differential Expression of LncRNAs Associated with Collagen in Mouse Placentas and Amniotic Sacs. Am J Reprod Immunol 2015;74:237-57. [Crossref] [PubMed]
- Barranco SC, Townsend CM Jr, Casartelli C, et al. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res 1983;43:1703-9. [PubMed]
- Lin C, Fu Z, Liu Y, et al. The establishment of human gastric carcinoma cell line (SGC7901). China Academic Journal Electronic Publishing House 1981;1:1-03.
- Akagi T, Kimoto T. Human cell line (HGC-27) derived from the metastatic lymph node of gastric cancer. Acta Med Okayama 1976;30:215-9. [PubMed]
- Naito Y, Kino I, Horiuchi K, et al. Promotion of collagen production by human fibroblasts with gastric cancer cells in vitro. Virchows Arch B Cell Pathol Incl Mol Pathol 1984;46:145-54. [Crossref] [PubMed]


