Comprehensive analysis of the NME gene family functions in breast cancer
Introduction
Breast cancer (BC) is the most prevalent female malignancy worldwide, with the highest morbidity and mortality rates (1). In 2019, approximately 268,600 new cases of invasive BC and 48,100 cases of ductal carcinoma in situ (DCIS) were reported in the female population of America. Furthermore, an estimated 41,760 females are predicted to die from BC (2). The clinical molecular subtypes of BC are based on the state of progesterone receptor (PR), estrogen receptor (ER), human epidermal growth factor receptor-2 (HER-2) and ki-67. The standard available treatments for BC include surgery, radiation, chemotherapy, hormone therapy, and targeted therapy (3), based on the particular BC molecular subtype. Although precision therapy can prolong the survival of BC patients, recurrence and metastasis remain the leading causes of death. Hence, the rapid identification of a reliable biomarker is required, which could facilitate the early diagnosis and prognosis in BC patients.
The human NME family of proteins was previously known as Nm23 proteins (4) or proteins “expressed in the non-metastatic cell.” It consists of 10 isoforms, named NME1-10, according to their subcellular localization and enzymatic activities. The conserved domain with nucleoside diphosphate kinase (NDPK) function is found in all NME family members, though not all are catalytically active (5). The human NME gene family is involved in multiple physiological and pathological activities, such as ciliary functions, proliferation, development, metastasis, and differentiation (6,7). Specifically, studies have revealed that nuclear translocation of NME1 and NME2 further modulate gene transcription via the DNA-binding abilities, which indicates potential co-regulator functions (5), despite the absence of canonical nuclear localization signal.
Additionally, NME1, NME5, NME7, and NME8 harbor the 3'–5' exonuclease activity (8), indicating their involvement in DNA recombination and repair. NME1 and NME3 may participate in repairing both single- and double-stranded breaks in DNA, contributing to the genomic instability and promoting malignant tumor progression. The subcellular localization of the NME gene family varies from the nucleus, cell membrane, mitochondria, and cytoplasm with different extents. Interestingly, extracellular NME protein has also been found in normal and tumor environments and consistently exhibits NDPK activity. Interestingly, the underlying mechanism which facilitates the nuclear translocation of NME proteins remains unclear due to the absence of the canonical nuclear localization signal. Indeed, clinical data also indicate that the NME gene family may be a critical prognostic indicator of several malignancies, such as gastric, neuroblastoma, and hematological malignancies, without definitive mechanisms. Traditionally, high levels of NME1 protein expression in melanoma, liver cancer, ovarian, lung, and BC are correlated with low metastatic potential, while the opposite phenomenon was observed in hematopoietic malignancies, neuroblastoma, and osteosarcoma. Additionally, the effects of NME1 on OS have not been established (7,9,10). Several therapeutic strategies using NME have been explored in tumor models over the past two decades, including re-expression of NME (11), gene therapy using NME delivery method (12,13), protein-based therapy to deliver cell-permeable NME protein (14), and lysophosphatidic acid (LPA) inhibitor (15).
The mechanism of Janus-faces of NME genes family oncoprotein interactions has not yet clarified (16). These extracellular NME proteins have previously been reported to show oncogenic functions or anti-differentiation capacities, which contradicts their known anti-metastatic function (17). The latest report has revealed that NME1 could enhance tumor growth and metastases in melanoma (18).
Clarifications on the precise functions of the NME gene family as oncogenes or tumor suppressor genes in BC patients are therefore critical. However, the diverse transcriptional expression, molecular functions, biological processes, and prognostic significance of most NME genes family require further elucidation in BC. In this study, a comprehensive analysis of the correlation of NME isoforms with the pathogenesis and progression of BC was performed via the incorporation of several widely-established databases, which could contribute to our current knowledge on the pathology, molecular significance and clinical predictive value in BC.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-1712).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Oncomine database
The transcriptional levels of NME in diverse types of malignancies were analyzed using the online cancer microarray database and data-mining platform (https://www.oncomine.org), ONCOMINE (19). ONCOMINE database contains 715 datasets and 86,733 samples covering most common cancer types. The mRNA expression of NME was compared between cancer and normal samples before determining the P values via a Students’ t-test. The P value and fold change (FC) were set at 0.01 and 2, respectively.
The Gene Expression Profiling Interactive Analysis (GEPIA) Dataset
GEPIA (http://gepia.cancer-pku.cn) is a newly developed web-based tool that facilitates expression analyses at the subtype level, based on TCGA and GTEx data (20). Gene and isoform expression can be analyzed by GEPIA, including general information, differential analysis, expression analysis, survival analysis, profiling plotting, similar genes detection, etc. Herein, the differential expression of NME genes in BRCA and normal samples was validated by the pathological stage. The P values via a Students’ t-test, and P<0.05 was considered statistically significant.
The Kaplan-Meier Plotter
Kaplan-Meier (KM) Plotter (www.kmplot.com) is a web-based tool used to assess the effect of various genes on survival across different cancers. It contained gene expression and survival data from the largest dataset on BC patients (21) and was employed to validate the prognostic value of the mRNA expression of the NME gene family. In order to evaluate the effects of NME gene expression on the OS of BC patients, samples were categorized into two groups based on the median gene expression (high versus low expression), followed by assessment using the KM survival plot, with a hazard ratio (HR) and corresponding 95% confidence intervals (CIs) as well as logrank P value. KM curves were plotted only on the JetSet best probe set of NMEs.
The cBioPortal for cancer genomics
The open-access and open-source cBioPortal (http://www.cbioportal.org/) for cancer genomics were developed at Memorial Sloan-Kettering Cancer Center (MSKCC) to explore multidimensional cancer genomics interactively (22,23). Functionally, it supports and stores data associated with DNA methylation, DNA copy-number, nonsynonymous mutations, enrichment, mRNA and miRNA expression, co-expression, and clinical parameters. The online cBioPortal tool rendered the calculation of the frequency of NME gene family alterations, including mutation, amplification, deletion, and mRNA expression z-scores (RNA Seq V2 RSEM). The heatmap demonstrated that the correlation of NME genes was procured by using the online tool (https://chart-studio.plot.ly). The counting data were expressed as rate (%). The correlation of the different genes was tested by Pearson’s correlation.
The STRING online database
The STRING database (https://string-db.org/) was used to search for interactions between known proteins and predictive proteins, followed by the construction of the protein-protein interaction (PPI) network on the NME gene family (24). The PPI network aimed to collect, score and integrate all publicly available sources of PPI information within various diseases, and complement these with computational predictions. The visualize PPI network determining the P values via a Students’ t-test. The P value and cut-off criteria of combined score were set at 0.05 and >0.4, respectively.
Functional enrichment and bioinformatics analysis
Metascape (http://metascape.org) is an online platform designed to provide a comprehensive gene list annotation and analysis resource. The Metascape database integrates over 40 independent bioinformatics bases, which could be utilized to extract abundant annotations, perform functional enrichment, interactome analysis, gene annotation, and establish PPI networks from lists of genes and proteins (25). Herein, the most frequently-changed related genes were identified via the analysis of gene lists containing the NME gene family by using Gene Ontology (GO) as well as the Kyoto Encyclopedia of Genes and Genomes (KEGG) tools within Metascape. Pearson’s correlation was determining the correlation. The correlation of the different genes was tested by Pearson’s correlation.
Results
The transcription expression of NME genes in BC patients
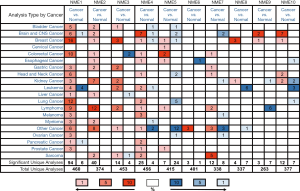
The identification of the NME gene family has been reported in the BC genome. ONCOMINE database analysis of NME transcriptional levels between tumor and normal samples in different cancer types (Figure 1) revealed upregulated mRNA expression of NME1, NME2, NME3, NME4, NME5, NME8, NME9, and NME10 in BC patients (Table 1). Moreover, the mRNA expression of NME1 increased from 2.006 to 3.309-fold in different types of BC compared to normal tissues (26-32). Curtis et al. revealed an upregulated mRNA expression of NME2 in Mucinous Breast Carcinoma (FC =2.004) (28). Moreover, the TCGA dataset showed that NME3 was elevated in Intraductal Cribriform Breast Adenocarcinoma, Invasive Ductal and Lobular Breast Carcinoma, and Mixed Lobular and Ductal Breast Carcinoma compared to the control samples, with FC of 2.783, 2.179 and 2.313 respectively. Meanwhile, they also demonstrated the overexpression of NME4 in Mucinous Breast Carcinoma (FC =2.190) and the overexpression of NME5 in Mucinous Breast Carcinoma (FC=2.493). According to the datasets from Ma Breast 4 and TCGA (30), NME8 expression was higher in tumor tissues than normal tissues as follows: (I) Ductal Breast Carcinoma in Situ Stroma (FC =2.640); (II) Invasive Ductal Breast Carcinoma (FC =2.434); (III) Invasive Lobular Breast Carcinoma (FC =2.017). Moreover, Finak et al. revealed the upregulated expression of NME9 (FC =3.482) in Invasive Breast Carcinoma Stroma than normal tissue (33). According to the dataset by Ma et al., NME10 was elevated in Invasive Ductal Breast Carcinoma (FC =2.018) (30). However, ONCOMINE analysis revealed no significant difference in mRNA levels of NME6 and NME7 between BC and normal samples.
Table 1
| Gene | Types of breast cancer vs. normal | Fold change | P value | t-test | Reference and/or Source |
|---|---|---|---|---|---|
| NME1 | Lobular Breast Carcinoma vs. Normal | 2.166 | 3.59E-4 | 5.080 | Sorlie T (26) |
| Ductal Breast Carcinoma vs. Normal | 2.826 | 5.98E-6 | 10.849 | Sorlie T (26) | |
| Lobular Breast Carcinoma vs. Normal | 2.303 | 0.003 | 4.567 | Perou CM (27) | |
| Ductal Breast Carcinoma vs. Normal | 3.309 | 6.05E-4 | 8.640 | Perou CM (27) | |
| Medullary Breast Carcinoma vs. Normal | 2.876 | 1.88E-17 | 13.844 | Curtis C (28) | |
| Mucinous Breast Carcinoma vs. Normal | 2.217 | 1.09E-20 | 12.957 | Curtis C (28) | |
| Tubular Breast Carcinoma vs. Normal | 2.021 | 3.23E-27 | 13.770 | Curtis C (28) | |
| invasive Ductal and Invasive Lobular Breast Carcinoma vs. Normal | 2.188 | 1.32E-29 | 14.086 | Curtis C (28) | |
| Breast Carcinoma vs. Normal | 2.101 | 3.99E-5 | 5.460 | Curtis C (28) | |
| Ductal Breast Carcinoma in Situ vs. Normal | 2.530 | 0.002 | 3.878 | Curtis C (28) | |
| Lobular Breast Carcinoma vs. Normal | 2.006 | 0.006 | 3.748 | Sorlie T (29) | |
| Ductal Breast Carcinoma vs. Normal | 2.841 | 1.25E-4 | 8.456 | Sorlie T (29) | |
| Invasive Ductal Breast Carcinoma Epithelia vs. Normal | 2.209 | 5.03E-6 | 5.959 | Ma XJ (30) | |
| Ductal Breast Carcinoma in Situ Epithelia vs. Normal | 2.527 | 0.004 | 3.275 | Ma XJ (30) | |
| Ductal Breast Carcinoma vs. Normal | 2.825 | 6.59E-11 | 8.351 | Richardson AL (31) | |
| Invasive Ductal Breast Carcinoma vs. Normal | 2.133 | 0.002 | 3.862 | Radvanyi L (32) | |
| NME2 | Mucinous Breast Carcinoma vs. Normal | 2.004 | 2.00E-11 | 8.537 | Curtis C (28) |
| NME3 | Intraductal Cribriform Breast Adenocarcinoma vs. Normal | 2.783 | 6.14E-36 | 26.856 | TCGA |
| Invasive Ductal and Lobular Breast Carcinoma vs. Normal | 2.179 | 1.76E-8 | 15.396 | TCGA | |
| Mixed Lobular and Ductal Breast Carcinoma vs. Normal | 2.313 | 2.01E-5 | 8.713 | TCGA | |
| NME4 | Mucinous Breast Carcinoma vs. Normal | 2.190 | 0.004 | 5.008 | TCGA |
| NME5 | Intraductal Cribriform Breast Adenocarcinoma vs. Normal | 2.493 | 2.58E-5 | 7.006 | TCGA |
| NME6 | NA | NA | NA | NA | NA |
| NME7 | NA | NA | NA | NA | NA |
| NME8 | Ductal Breast Carcinoma in Situ Stroma vs. Normal | 2.640 | 0.002 | 3.45 | Ma XJ (30) |
| Invasive Ductal Breast Carcinoma Stroma vs. Normal | 2.434 | 0.008 | 2.958 | Ma XJ (30) | |
| Invasive Lobular Breast Carcinoma vs. Normal | 2.017 | 6.27E-8 | 6.022 | TCGA | |
| NME9 | Invasive Breast Carcinoma Stroma vs. Normal | 3.84 | 2.52E-15 | 13.022 | Finak G (33) |
| NME10 | Invasive Ductal Breast Carcinoma vs. Normal | 2.018 | 0.003 | 3.120 | Ma XJ (30) |
NA, not available.
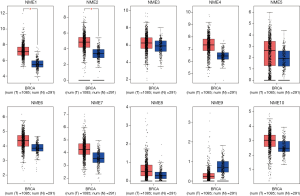
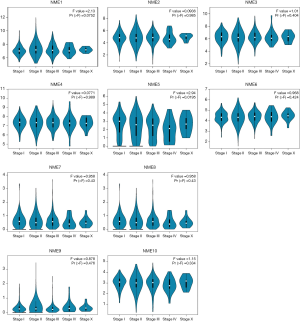
The GEPIA dataset was subsequently adopted to compare NME mRNA expression between BC and normal samples. The expression of NME1 and NME2 was significantly elevated in BC tissues compared to normal specimens (P<0.05), while NME3-10 expression was not statistically significant (Figure 2). Additionally, the expression levels of NME1-10 were not significantly different among different stages of BC (Figure 3).
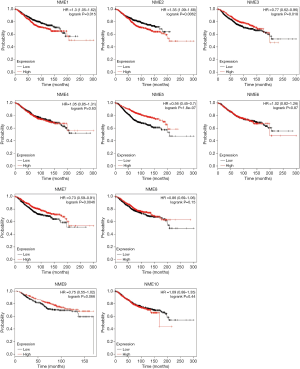
Furthermore, the KM Plotter was employed to study the effects of NME genes in BC survival. As a result, upregulated NME1 and NME2 levels were significantly associated with poor OS (P<0.05) in BC patients (Figure 4). Hence, the overexpression of NME1 and NME2 potentially indicates poor prognosis in BC patients, while NME3, NME5, and NME7 displayed effects opposite to NME1 and NME2.
Genetic alteration and correlation of NME genes in BC
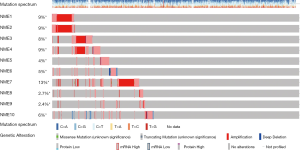
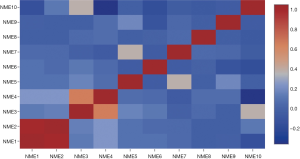
The cBioportal online tool and analysis of the TCGA database revealed NME genetic alterations, followed by the identification of any genetic associations. Consequently, genetic alterations of NME genes were detected in 450/1,098 (41%) of BC samples (Figure 5), including mRNA and protein expression, mutation, amplification as well as homozygous deletion. The cBioPortal was additionally adopted to investigate the interactions between specific NME genes (including Pearson’s correlation) (Figure 6).
Predicted functions and pathway enrichment analysis of NME genes in BC
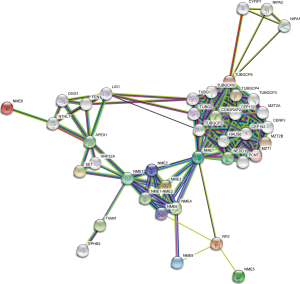
The PPI network for the NMEs gene family revealed that the 30 most frequently changed neighboring genes (Figure 7). A list of the expressed NME genes was compiled along with the most frequently changed associated genes before GO and KEGG analysis using Metascape (Figure 8). NME gene alterations influenced the following processes: R-HAS-380270: Recruitment of mitotic centrosome and complexes; GO:0006228: UTP biosynthetic process; R-HAS-380259: Loss of NlP from mitotic centrosomes; hsa03410: Base excision repair; CORUM:3714: Pericentrin-GCP complex; GO:1902850: microtubule cytoskeleton organization involved in mitosis; GO:0045454: cell redox homeostasis; GO:0031346: positive regulation of cell projection organization.
Discussion
Continuous advances in precision treatment have prolonged the survival of BC patients and enhanced their quality of life. However, tumor recurrence and metastasis are still the main causes of death in BC patients. The underlying mechanisms remain poorly understood, highlighting a critical need to explore and identify more efficient molecular markers for the early diagnosis and treatment of BC. Currently, the significance of NME gene family expression in the initiation and prognosis of BC remains largely unknown. The NME gene family display NDPK functions that share DNA binding, DNA damage repair, class switch recombination, and transcription (34), highlighting their potential in tumor progression (5). Essentially, the NME gene family is involved in multiple functions across three major cellular processes, namely protein histidine phosphorylation, nucleotide channeling and membrane remodeling, tumor progression, and metastasis (35).
Our study showed alterations of NME genes in ~41% (450/1,098) of BC specimens in the form of increased mRNA levels, deep deletion, or amplification. We also revealed that increased mRNA expression of NME1 and NME2 was significantly correlated with poor prognosis in BC patients. Amongst the NME genes family, NME1 and NME2 are the most commonly investigated in BC. Several types of research have demonstrated that NME1 not only suppresses metastasis, but enhances cancer cell growth and metastatic properties, including melanoma (18), BC (36), lung adenocarcinoma (37), and neuroblastoma (38). In agreement with the previous study, NME1 is characterized by the capacity to inhibit the metastatic phenotype of tumor cells, without affecting primary tumor growth (5,16). This positive correlation in NME1 and NME2 is attributed to the 88% sequential homology, as well as the similar functional and structural attributes. Conery et al. revealed that the loss of NME1, suspected to enhance metastasis, might also trigger the acquisition of chromosomal instability at an earlier stage of tumor development by in vitro assay in HBE125 E6/E7 and 293T cells (39). Meanwhile, another study confirmed that NME1, as a novel nonhomologous end-joining (NHEJ) factor, promoted genome stability (40).
It is worth noting that high expression of NME3, NME5, and NME7 was correlated with long-term survival, despite contradictory findings in transcriptional expression between ONCOMINE and GEPIA dataset. Similarly, one study confirmed NME3 as a positive downstream regulator of flagellin-mediated NFκB signaling, which is highly correlated with the expression of toll-like receptor 5 (TLR5) in ovarian, lung and BCs, and increased OS. Interestingly, they further recognized the proinflammatory effects of NME3, a signaling downstream target of TLR5, which might facilitate tumor immunotherapies (41). By using expression microarrays and array comparative genomic hybridization, Parris et al. have revealed 97 invasive diploid breast tumors with changes in DNA copy number and transcriptional expression. The integrative analysis also showed that downregulated NME5 and other genes play vital roles in BC carcinogenesis and progression, highlighting their potential as novel therapeutic targets (42). NME6 and NME7 are critically involved in the renewal of embryonic stem cells and oncogenesis (43). NME7, a poorly characterized member of the NME family, is a component of the γ-tubulin ring complex (γTuRC) and regulates the microtubule-nucleating activity (44).
Although a small number of studies have suggested that NME4, NME8, NME9, and NME10 are highly expressed in BC; the comprehensive analysis undertaken in our study showed no correlation with clinical diagnosis and prognosis. NME4, also known as a moonlighting protein, is transported to the mitochondria through the mitochondria-specific targeting signal. Unlike the other NME family members, NME4 requires cleavage for its catalytic activity (45). NME8 also termed TXNDC3, nm23-h8, sptrx-2, SPTRX2, hels-99, and CILD6 is currently studied in Alzheimer’s disease and osteoarthritis (46,47). NME9, also known as Thioredoxin Domain-Containing Protein 6 (TXDC6), is a novel eutherian NME8-related protein (4), and NME10, recognized as retinitis pigmentosa 2 (RP2), is the outgroup member of the NME family. Current research has reported that RP2 mutation is involved with nearly 20% X-linked retinitis pigmentosa. Moreover, RP2 in a structurally compromised state has also been confirmed as the predominating pathogenic consequence for the majority of RP2 mutations. The C-terminal domain of RP2, however, can maintain the overall protein stability (48).
In this study, we constructed and explored the PPI network for the NMEs family. Furthermore, the association of NME gene expression and the most frequently changed correlated genes with BC carcinogenesis and prognosis was assessed by GO and KEGG analysis. The gene families with the highest frequencies were NIPA, TUBG, TUBGCP, etc. As a result, we conclude that the following pathways should be further investigated, including R-HAS-380270: Recruitment of mitotic centrosome and complexes; GO:0006228: UTP biosynthetic process; R-HAS-380259: Loss of NlP from mitotic centrosomes; hsa03410: Base excision repair; and CORUM:3714: Pericentrin-GCP complex.
Collectively, our research suggested that the elevated expression of NME1 and NME2 could potentially act as a molecular marker for the identification of BC patients with poor prognosis, while NME3, NME5, and NME7 may still play tumor-suppressive roles. Furthermore, NME1 and NME2 are likely to be a strong prognostic biomarker and potential therapeutic target to enhance the diagnosis and therapy of BC. Moreover, large-scale studies are needed to validate the functions of NME3, NME5, and NME7.
Acknowledgments
We also acknowledge the contributions of ONCOMINE, GEPIA, Kaplan-Meier plotter, cBioPortal, String as well as metascape databases.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1712
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1712). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ahmad A. Breast Cancer Statistics: Recent Trends. Adv Exp Med Biol 2019;1152:1-7. [Crossref] [PubMed]
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. [Crossref] [PubMed]
- Harbeck N, Gnant M. Breast cancer. Lancet 2017;389:1134-50. [Crossref] [PubMed]
- Desvignes T, Pontarotti P, Fauvel C, et al. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol 2009;9:256. [Crossref] [PubMed]
- Puts GS, Leonard MK, Pamidimukkala NV, et al. Nuclear functions of NME proteins. Lab Invest 2018;98:211-8. [Crossref] [PubMed]
- Boissan M, Dabernat S, Peuchant E, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem 2009;329:51-62. [Crossref] [PubMed]
- Marino N, Nakayama J, Collins JW, et al. Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metastasis Rev 2012;31:593-603. [Crossref] [PubMed]
- Zhang Q, McCorkle JR, Novak M, et al. Metastasis suppressor function of NM23-H1 requires its 3'-5' exonuclease activity. Int J Cancer 2011;128:40-50. [Crossref] [PubMed]
- Snider NT, Altshuler PJ, Omary MB. Modulation of cytoskeletal dynamics by mammalian nucleoside diphosphate kinase (NDPK) proteins. Naunyn Schmiedebergs Arch Pharmacol 2015;388:189-97. [Crossref] [PubMed]
- McCorkle JR, Leonard MK, Kraner SD, et al. The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genomics Proteomics 2014;11:175-94. [PubMed]
- Palmieri D, Halverson DO, Ouatas T, et al. Medroxyprogesterone acetate elevation of Nm23-H1 metastasis suppressor expression in hormone receptor-negative breast cancer. J Natl Cancer Inst 2005;97:632-42. [Crossref] [PubMed]
- Li J, Zhou J, Chen G, et al. Inhibition of ovarian cancer metastasis by adeno-associated virus-mediated gene transfer of nm23H1 in an orthotopic implantation model. Cancer Gene Ther 2006;13:266-72. [Crossref] [PubMed]
- Li Z, Xiang J, Zhang W, et al. Nanoparticle delivery of anti-metastatic NM23-H1 gene improves chemotherapy in a mouse tumor model. Cancer Gene Ther 2009;16:423-9. [Crossref] [PubMed]
- Lim J, Jang G, Kang S, et al. Cell-permeable NM23 blocks the maintenance and progression of established pulmonary metastasis. Cancer Res 2011;71:7216-25. [Crossref] [PubMed]
- Marshall JC, Collins JW, Nakayama J, et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. J Natl Cancer Inst 2012;104:1306-19. [Crossref] [PubMed]
- Vlatkovic N, Chang SH, Boyd MT. Janus-faces of NME-oncoprotein interactions. Naunyn Schmiedebergs Arch Pharmacol 2015;388:175-87. [Crossref] [PubMed]
- Romani P, Ignesti M, Gargiulo G, et al. Extracellular NME proteins: a player or a bystander? Lab Invest 2018;98:248-57. [Crossref] [PubMed]
- Wang Y, Leonard MK, Snyder DE, et al. NME1 Drives Expansion of Melanoma Cells with Enhanced Tumor Growth and Metastatic Properties. Mol Cancer Res 2019;17:1665-74. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-D13. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003;100:8418-23. [Crossref] [PubMed]
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74. [Crossref] [PubMed]
- Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009;11:R7. [Crossref] [PubMed]
- Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121-32. [Crossref] [PubMed]
- Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A 2005;102:11005-10. [Crossref] [PubMed]
- Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008;14:518-27. [Crossref] [PubMed]
- Zheng S, Kusnadi A, Choi JE, et al. NME proteins regulate class switch recombination. FEBS Lett 2019;593:80-7. [Crossref] [PubMed]
- Boissan M, Schlattner U, Lacombe ML. The NDPK/NME superfamily: state of the art. Lab Invest 2018;98:164-74. [Crossref] [PubMed]
- Iqbal B, Masood A, Lone MM, et al. Polymorphism of Metastasis Suppressor Genes MKK4 and NME1 in Kashmiri Patients with Breast Cancer. Breast J 2016;22:673-7. [Crossref] [PubMed]
- Yang M, Sun Y, Sun J, et al. Differentially expressed and survival-related proteins of lung adenocarcinoma with bone metastasis. Cancer Med 2018;7:1081-92. [Crossref] [PubMed]
- Tan CY. CL. C. NDPKA is not just a metastasis suppressor - be aware of its metastasis-promoting role in neuroblastoma. Lab Invest 2018;98:219-27. [Crossref] [PubMed]
- Conery AR, Sever S, Harlow E. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proc Natl Acad Sci U S A 2010;107:15461-6. [Crossref] [PubMed]
- Xue R, Peng Y, Han B, et al. Metastasis suppressor NME1 promotes non-homologous end joining of DNA double-strand breaks. DNA Repair (Amst) 2019;77:27-35. [Crossref] [PubMed]
- Flentie K, Gonzalez C, Kocher B, et al. Nucleoside Diphosphate Kinase-3 (NME3) Enhances TLR5-Induced NFkappaB Activation. Mol Cancer Res 2018;16:986-99. [Crossref] [PubMed]
- Parris TZ, Danielsson A, Nemes S, et al. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin Cancer Res 2010;16:3860-74. [Crossref] [PubMed]
- Wang CH, Ma N, Lin YT, et al. A shRNA functional screen reveals Nme6 and Nme7 are crucial for embryonic stem cell renewal. Stem Cells 2012;30:2199-211. [Crossref] [PubMed]
- Liu P, Choi YK, Qi RZ. NME7 is a functional component of the gamma-tubulin ring complex. Mol Biol Cell 2014;25:2017-25. [Crossref] [PubMed]
- Lacombe ML, Tokarska-Schlattner M, Boissan M, et al. The mitochondrial nucleoside diphosphate kinase (NDPK-D/NME4), a moonlighting protein for cell homeostasis. Lab Invest 2018;98:582-8. [Crossref] [PubMed]
- Kim JH. Genetics of Alzheimer's Disease. Dement Neurocogn Disord 2018;17:131-6. [Crossref] [PubMed]
- Shi D, Nakamura T, Nakajima M, et al. Association of single-nucleotide polymorphisms in RHOB and TXNDC3 with knee osteoarthritis susceptibility: two case-control studies in East Asian populations and a meta-analysis. Arthritis Res Ther 2008;10:R54. [Crossref] [PubMed]
- Liu F, Qin Y, Yu S, et al. Pathogenic mutations in retinitis pigmentosa 2 predominantly result in loss of RP2 protein stability in humans and zebrafish. J Biol Chem 2017;292:6225-39. [Crossref] [PubMed]