
KIF4A is a promising prognostic marker and correlates with immune infiltration in clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC), originates from the renal tubular epithelium, is one of the most common malignant tumors of the urinary system. Statistics show that its incidence has been increasing by 2% annually (1). Clear cell renal cell carcinoma (ccRCC) is the most common pathological subtype with high mortality and recurrence rates, accounting for 70–85% of all RCC cases (2). Although considerable efforts have been made to improve the treatment of this cancer, the prognosis of ccRCC remains unsatisfactory, especially for patients with recurrent or metastatic tumors (3-5). Therefore, it is critically urgent to find effective biomarkers for early diagnosis and treatment of ccRCC.
Kinesins encode a protein that is microtubule-dependent molecular motor that participates in essential cellular functions such as intracellular transport and cell division (6,7). Numerous studies have revealed that kinesins play vital roles in the occurrence and development of various types of malignancies (8-11). Currently, the effect of kinesins on the progression of cancers has mainly focused on its role in tumor cell proliferation, and therefore immune infiltration in the tumor microenvironment is less known. Some members of the kinesin family were reported to regulate the tumor microenvironment. A recent study demonstrated that kinesin family member 18A (KIF18A) regulated the differentiation and activation of dendritic cells (12). Elsewhere, the kinesin family member 20A (KIF20A) showed the potential to be an immunotherapeutic effect in glioma (13).
KIF4A is a member of the kinesin family and correlated with poor prognosis and progression of multiple human cancers (14-17). However, the expression and prognostic value of KIF4A in ccRCC and its correlation with tumor-infiltrating immune cells have not been reported so far.
In this study, we used the publicly available online database to analyze the expression level of KIF4A, prognostic value, and its correlation with clinicopathological characteristics and tumor-infiltrating immune cells in ccRCC.
Methods
TIMER database analysis
The Tumor Immunity Resource (TIMER) (https://cistrome.shinyapps.io/timer/) database contains mRNA sequencing (mRNA-seq) data for 10,897 samples of 32 human cancers from the Cancer Genome Atlas (TCGA) (18). The mRNA expression of KIF4A in different types of human tumors was analyzed using TIMER database. The TIMER database is an ideal tool for systematic analysis of immune infiltration in various cancer types (18). We assessed the association of KIF4A expression with the abundance of different types of immune infiltrating cells. The relationship between KIF4A expression level and tumor purity was analyzed. In addition, correlations between KIF4A expression and various immune markers were further investigated via TIMER database.
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu/) includes TCGA grade 3 RNA-seq and clinical data of 31 human cancer types. The platform can be used to analyze the relationship between gene expression and individual cancer stages, tumor grade or other clinicopathological features (19). UALCAN was utilized to determine the correlation between KIF4A expression and ccRCC clinicopathological features.
GEPIA database analysis
The Gene Expression Profiling Analysis Interaction Analysis (GEPIA) (http://gepia.cancer-pku.cn) is an interactive web that includes 9,736 tumors and 8,587 normal samples from TCGA and the Genotype-Tissue Expression (GTEx) projects (20). GEPIA was used to analyze the association of KIF4A expression with the survival rate of ccRCC patients. The 511 ccRCC patients with available overall survival (OS) and disease-free survival (DFS) time data were divided into low- and high-expression groups by the median transcripts per kilobase million (TPM), a logarithmic rank P<0.05 was considered significant.
Statistical analysis
Wilcoxon test was used for differential expression analysis. Survival analysis was displayed using Kaplan-Meier curves, with statistical significance evaluated using the Log-rank test. Spearman correlation coefficient was applied to assess correlations between KIF4A and immune infiltration level. P<0.05 was considered statistically significant.
Results
The KIF4A mRNA expression profile in different human cancer types
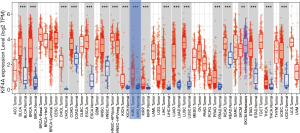
To determine the expression of KIF4A between tumorigenic and noncancerous tissues, the mRNA expression levels of KIF4A in multiple cancer types and healthy control samples were analyzed using the TIMER database for various tumors. The results showed that KIF4A mRNA was highly expressed in various cancer types compared with adjacent normal tissues, including kidney renal clear cell carcinoma (KIRC), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (Figure 1).
Relationship between KIF4A expression and clinicopathological features of ccRCC patients
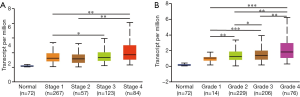
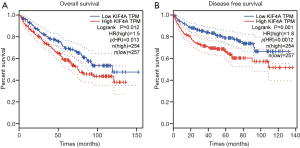
By using the UALCAN database, the relationship between KIF4A expression and clinicopathological features of patients with ccRCC was determined. The results showed that high expression of KIF4A was positively correlated with the clinical stage and histological grade (Figure 2A,B), indicating that ccRCC patients with high KIF4A expression levels had poor clinical outcome. In addition, we performed Kaplan-Meier analyses for OS and DFS to evaluate the prognostic value of KIF4A in ccRCC patients. Results showed that increased KIF4A mRNA expression was correlated with shorter OS (HR =1.5, log-rank P=0.012) and lower DFS (HR =1.8, log-rank P=0.001) of ccRCC patients (Figure 3A,B). The results demonstrated that high expression of KIF4A might predict poor prognosis of ccRCC.
Correlation between KIF4A expression with tumor purity and immune cells infiltration levels
By using the Timer database, we analyzed the correlation between KIF4A expression with tumor purity and immune cells (including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) infiltration level in ccRCC (Figure 4). The results showed that KIF4A expression was negatively correlated with tumor purity in ccRCC. Moreover, KIF4A expression was positively correlated with infiltration of B cells (r=0.282, P=7.92e-10), CD4+ T (r=0.280, P=9.86e-10) and CD8+ T cells (r=0.237, P=5.19e-07), macrophages (r=0.263, P=1.64e-08), neutrophils (r=0.376, P=7.28e-17), and dendritic cells (r=0.424, P=3.07e-21). These findings reveal that KIF4A plays a crucial role in immune infiltration in the KIRC microenvironment.
Correlation between KIF4A expression and immune markers
To further explore the relationship between KIF4A and different types of immune infiltrating cells, we analyzed the correlation between KIF4A expression and the immune markers of diverse immune infiltrating cells using TIMER database, including CD8+ T cells, B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, NK cells and DCs in KIRC, as well as the various functional T cells, such as Th1 cells, Th2 cells, Tfh cells, Th17 cells, and Tregs, exhausted T cells. Given that immune infiltration can be affected by tumor purity, the analysis was performed after correcting for tumor purity. We found that the expression level of KIF4A was significantly associated with most immune markers of immune cells and different T cells in KIRC (Table 1). Specifically, high KIF4A expression was significantly correlated with high levels of multiple immune markers in KIRC, such as CD8+ T cell markers, CD8A (r=0.251; P=4.71e-08), and CD8B (r=0.204; P=1.06e-05); B cell markers, CD19 (r=0.230; P=5.92e-07), and CD79A (r=0.165; P=3.83e-04); Monocyte markers, CD86 (r=0.332; P=2.69e-13), and CSF1R (r=0.314; P=5.32e-12); TAM markers, CD68 (r=0.336; P=1.15e-13), and IL10 (r=0.317; P=3.41e-12); M1 Macrophage markers NOS2 (r=0.137; P=3.14e-03), IRF5 (r=0.144; P=1.92e-03), and PTGS2 (r=0.191; P=3.71e-05); M2 Macrophage markers CD163 (r=0.365; P=5.31e-16), VSIG4 (r=0.319; P=2.16e-12), and MS4A4A (r=0.331; P=2.81e-13); Neutrophils markers ITGAM (r=0.296; P=8.79e-11), and CCR7 (r=0.294; P=1.13e-10); Natural killer cell marker KIR2DL4 (r=0.166; P=3.46e-04); Dendritic cell markers HLA-DPB1 (r=0.197; P=2.01E-05), HLA-DRA (r=0.233; P=4.34E-07), HLA-DPA1 (r=0.243; P=1.22E-07), CD1C (r=0.101; P=3.03E-02), NRP1 (r=0.227; P=8.56E-07), and ITGAX (r=0.229; P=7.07E-07); Th1 markers, TBX21 (r=0.137; P=3.14E-03), IFNG (r=0.283; P=6.15E-10), TNF (r=0.17; P=1.61E-04), STAT1 (r=0.424; P=1.63e-21), and STAT4 (r=0.325; P=8.31e-13); Th2 marker STAT5A (r=0.232; P=4.58E-07); Tfh marker BCL6 (r=0.195; P=2.57E-05); Th17 marker STAT3 (r=0.288; P=2.96E-10); Treg markers, FOXP3 (r=0.367; P=4.01e-16), CCR8 (r=0.384; P=1.22e-17), and TGFB1 (r=0.363; P=8.63e-16); and T cell exhaustion markers, PDCD1 (r=0.253; P=3.82E-08), CTLA4 (r=0.240; P=1.93E-07), LAG3 (r=0.284; P=5.69E-10), and GZMB (r=0.138; P=2.95E-03). These findings further suggest that KIF4A expression correlates with infiltration of immune cells in the KIRC microenvironment.
Table 1
| Description | Gene markers | KIF4A | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P | Cor | P | |||
| CD8+ T cell | CD8A | 0.268 | **** | 0.251 | **** | |
| CD8B | 0.222 | **** | 0.204 | **** | ||
| B cell | CD19 | 0.254 | **** | 0.230 | **** | |
| CD79A | 0.187 | **** | 0.165 | *** | ||
| Monocyte | CD86 | 0.347 | **** | 0.332 | **** | |
| CSF1R | 0.343 | **** | 0.314 | **** | ||
| TAM | CCL2 | −0.021 | 0.632 | −0.054 | 0.245 | |
| CD68 | 0.336 | **** | 0.336 | **** | ||
| IL10 | 0.335 | **** | 0.317 | **** | ||
| M1 macrophage | NOS2 | 0.170 | **** | 0.137 | ** | |
| IRF5 | 0.143 | *** | 0.144 | ** | ||
| PTGS2 | 0.209 | **** | 0.191 | **** | ||
| M2 macrophage | CD163 | 0.381 | **** | 0.365 | **** | |
| VSIG4 | 0.353 | **** | 0.319 | **** | ||
| MS4A4A | 0.349 | **** | 0.331 | **** | ||
| Neutrophils | CEACAM8 | 0.017 | 0.695 | 0.048 | 0.300 | |
| ITGAM | 0.325 | **** | 0.296 | **** | ||
| CCR7 | 0.312 | **** | 0.294 | **** | ||
| Natural killer cell | KIR2DL1 | 0.036 | 0.401 | 0.033 | 0.481 | |
| KIR2DL3 | 0.028 | 0.519 | 0.012 | 0.789 | ||
| KIR2DL4 | 0.174 | **** | 0.166 | *** | ||
| KIR3DL1 | −0.002 | 0.959 | 0.010 | 0.834 | ||
| KIR3DL2 | 0.017 | 0.693 | 0.016 | 0.732 | ||
| KIR3DL3 | 0.083 | 0.055 | 0.081 | 0.084 | ||
| KIR2DS4 | 0.007 | 0.872 | 0.007 | 0.884 | ||
| Dendritic cell | HLA-DPB1 | 0.214 | **** | 0.197 | **** | |
| HLA-DQB1 | 0.116 | ** | 0.097 | * | ||
| HLA-DRA | 0.245 | **** | 0.233 | **** | ||
| HLA-DPA1 | 0.253 | **** | 0.243 | **** | ||
| CD1C | 0.131 | ** | 0.101 | * | ||
| NRP1 | 0.257 | **** | 0.227 | **** | ||
| ITGAX | 0.230 | **** | 0.229 | **** | ||
| Th1 | TBX21 | 0.158 | *** | 0.137 | ** | |
| STAT4 | 0.336 | **** | 0.325 | **** | ||
| STAT1 | 0.422 | **** | 0.424 | **** | ||
| IFNG | 0.293 | **** | 0.283 | **** | ||
| TNF | 0.185 | **** | 0.175 | *** | ||
| Th2 | GATA3 | 0.085 | * | 0.064 | 0.170 | |
| STAT6 | 0.048 | 0.273 | 0.060 | 0.197 | ||
| STAT5A | 0.278 | **** | 0.232 | **** | ||
| IL13 | 0.024 | 0.575 | 0.032 | 0.487 | ||
| Tfh | BCL6 | 0.193 | **** | 0.195 | **** | |
| Th17 | STAT3 | 0.329 | **** | 0.288 | **** | |
| IL17A | 0.050 | 0.249 | 0.039 | 0.405 | ||
| Treg | FOXP3 | 0.377 | **** | 0.367 | **** | |
| CCR8 | 0.388 | **** | 0.384 | **** | ||
| STAT5B | 0.062 | 0.153 | 0.051 | 0.279 | ||
| TGFB1 | 0.419 | **** | 0.363 | **** | ||
| T cell exhaustion | PDCD1 | 0.256 | **** | 0.253 | **** | |
| CTLA4 | 0.257 | **** | 0.240 | **** | ||
| LAG3 | 0.308 | **** | 0.284 | **** | ||
| HAVCR2 | 0.108 | * | 0.085 | 0.069 | ||
| GZMB | 0.166 | *** | 0.138 | ** | ||
*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. KIF4A, kinesin family member 4A; ccRCC, clear cell renal cell carcinoma; TIMER, Tumor Immune Estimation Resource; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, Follicular helper T cell; Treg, regulatory T cell; Cor, R value of Spearman’s correlation; None, correlation without adjustment; Purity, correlation adjusted by purity.
Discussion
ccRCC is one of the most common malignant tumors of the urinary system, though advances in the development of strategies for treating ccRCC, the prognosis of the disease has remained poor, especially for patients with recurrent or metastatic tumors. It is urgent to find novel diagnostic and therapeutic targets of ccRCC and investigate the potential mechanisms.
Previous studies have demonstrated that KIF4A regulates multiple cellular processes, such as intracellular transport and cell division (21,22). Accumulating evidence indicates that KIF4A is an oncogene in various malignant tumors. For instance, KIF4A led to the induction of p21-mediated cell cycle progression, thereby facilitating the proliferation of colorectal cancer cells (14). In hepatocellular carcinoma, KIF4A exerts oncogenic effects by promoting the proliferation and invasion of tumor cells (15). In addition, KIF4A was found to be upregulated and correlated with poor clinical prognosis in prostate cancer (16). In lung adenocarcinoma cells, KIF4A contributes to chemotherapeutic drug resistance by transporting LRP-based vaults along the microtubules towards the cell membrane (17). Notably, these studies are mostly concerned with the theory of cell division. However, available data suggest that some kinesin family members have been found to regulate immune infiltration in the tumor microenvironment, such as KIF18A and KIF20A (12,13). In this study, we analyzed the expression and prognostic value of KIF4A in ccRCC. The correlation between KIF4A expression with tumor-infiltrating immune cells was also determined.
Here, we confirmed that KIF4A was significantly upregulated in multiple tumor types, and higher expression of KIF4A was closely associated with advanced clinical stage, high histological grade, and poor prognosis of ccRCC patients. Tumor-infiltrating lymphocytes are independent predictors of sentinel lymph node status and survival in cancers. Thus, we investigated whether the expression of KIF4A is related to the immune infiltration levels in ccRCC. Results showed that KIF4A expression was significantly associated with tumor purity and infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells in ccRCC. These findings suggest that KIF4A plays a specific role in the immune infiltration cells of ccRCC, especially for neutrophils and DCs. Furthermore, we found that KIF4A expression was significantly associated with most immune markers in ccRCC. For example, gene markers of TAM such as CD68, and IL10; gene markers of M1 macrophage such as NOS2, IRF5, and PTGS2; and gene markers of M2 macrophage such as CD163, VSIG4, and MS4A4A were significant correlated with KIF4A expression, showing the potential of KIF4A to regulate macrophage polarization in ccRCC. We also found that Th1 markers, TBX21, IFNG, TNF, STAT1, and STAT4; Th2 marker STAT5A; Tfh marker BCL6; Th17 marker STAT3; and Treg markers, FOXP3, CCR8, and TGFB1 were positively correlated with KIF4A expression. This indicates the role of KIF4A in regulating tumor infiltration of T helper cells. In addition, KIF4A expression in ccRCC was positively correlated with T cell exhaustion markers, such as PDCD1 and CTLA4, they are the key inhibitory immune checkpoint proteins that overexpressed in different human cancers helps tumor cells to evade immune responses by suppressing T cell function, thereby promoting cancer progression (23,24).
The current study advances our understanding of the relationship between KIF4A and ccRCC. Nonetheless, there are several limitations in this study. Firstly, our study was performed based on data from several public databases (TIMER, UALCAN and GEPIA database), which has not been further verified in experiments. Secondly, we failed to clearly evaluate the protein levels of KIF4A in ccRCC from the online database. Finally, the specific mechanisms responsible for immune infiltration in ccRCC needed further exploration.
In conclusion, our results show that KIF4A mRNA is significantly higher in different human cancers than in normal control tissues. Moreover, high expression of KIF4A correlates with poor clinical outcomes in ccRCC patients and high immune infiltration levels in KIRC. KIF4A is a promising prognostic marker and correlates with immune infiltration in ccRCC. Nevertheless, further molecular and cellular experimental evidence is required to validate our conclusions.
Acknowledgments
Funding: This project was supported and funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1937). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The public databases mentioned in this study are publicly available for re-analyzing, and no ethical approval was required by the local ethics committees, so that this study does not require the ethics approval.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Fernández-Pello S, Hofmann F, Tahbaz R, et al. A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma. Eur Urol 2017;71:426-36. [Crossref] [PubMed]
- Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol 2015;67:740-9. [Crossref] [PubMed]
- Barata PC, Rini BI. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J Clin 2017;67:507-24. [Crossref] [PubMed]
- Myers SM, Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med Chem 2016;8:463-89. [Crossref] [PubMed]
- Mann BJ, Wadsworth P. Kinesin-5 Regulation and Function in Mitosis. Trends Cell Biol 2019;29:66-79. [Crossref] [PubMed]
- Song X, Zhang T, Wang X, et al. Distinct Diagnostic and Prognostic Values of Kinesin Family Member Genes Expression in Patients with Breast Cancer. Med Sci Monit 2018;24:9442-64. [Crossref] [PubMed]
- Xiao KH, Teng K, Ye YL, et al. Kinesin family member C1 accelerates bladder cancer cell proliferation and induces epithelial-mesenchymal transition via Akt/GSK3β signaling. Cancer Sci 2019;110:2822-33. [Crossref] [PubMed]
- Yan H, Zhu C, Zhang L. Kinesin family member 18B: A contributor and facilitator in the proliferation and metastasis of cutaneous melanoma. J Biochem Mol Toxicol 2019;33:e22409. [Crossref] [PubMed]
- Li H, Zhang W, Sun X, et al. Overexpression of kinesin family member 20A is associated with unfavorable clinical outcome and tumor progression in epithelial ovarian cancer. Cancer Manag Res 2018;10:3433-50. [Crossref] [PubMed]
- Kim S, Cho YB, Song CU, et al. Kinesin family member KIF18A is a critical cellular factor that regulates the differentiation and activation of dendritic cells. Genes Genomics 2020;42:41-6. [Crossref] [PubMed]
- Saito K, Ohta S, Kawakami Y, et al. Functional analysis of KIF20A, a potential immunotherapeutic target for glioma. J Neurooncol 2017;132:63-74. [Crossref] [PubMed]
- Hou PF, Jiang T, Chen F, et al. KIF4A facilitates cell proliferation via induction of p21-mediated cell cycle progression and promotes metastasis in colorectal cancer. Cell Death Dis 2018;9:477. [Crossref] [PubMed]
- Hou G, Dong C, Dong Z, et al. Upregulate KIF4A Enhances Proliferation, Invasion of Hepatocellular Carcinoma and Indicates poor prognosis Across Human Cancer Types. Sci Rep 2017;7:4148. [Crossref] [PubMed]
- Gao H, Chen X, Cai Q, et al. Increased KIF4A expression is a potential prognostic factor in prostate cancer. Oncol Lett 2018;15:7941-7. [Crossref] [PubMed]
- Pan LN, Zhang Y, Zhu CJ, et al. Kinesin KIF4A is associated with chemotherapeutic drug resistance by regulating intracellular trafficking of lung resistance-related protein. J Zhejiang Univ Sci B 2017;18:1046-54. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-e110. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer 2012;12:527-39. [Crossref] [PubMed]
- Sheng L, Hao SL, Yang WX, et al. The multiple functions of kinesin-4 family motor protein KIF4 and its clinical potential. Gene 2018;678:90-9. [Crossref] [PubMed]
- Marin-Acevedo JA, Dholaria B, Soyano AE, et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol 2018;11:39. [Crossref] [PubMed]
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450-61. [Crossref] [PubMed]



