
Expression profiling analysis reveals molecular mechanism of Lnc00675 downregulation promoting cell apoptosis in acute myeloid leukemia U937 cells
Introduction
Acute myeloid leukemia (AML), an aggressive malignancy with poor prognosis, is the most common in adult leukemia. The therapeutic efficacy of patients with AML remains poor, with only 40% young patients (<60 years old) or 10% patients (>60 years old) achieving long-term survival (1). The pathological mechanism of AML is a series of events including changes in cell proliferation, differentiation, and apoptosis caused by pathogenic factors, such as somatic mutations, cytogenetic abnormalities, epigenetic changes (2,3).
Long non-coding RNAs (lncRNAs) are e a class of RNAs longer than 200 nucleotides, which don’t have the function of encoding proteins (4). LncRNA could affect protein-coding gene regulation, cell proliferation and apoptosis, tumor cell resistance to radio- and chemotherapy and pathological processes by participating in transcriptional regulation and post-transcriptional regulation (5-8). Accumulating evidence supports that misregulation of lncRNA-based epigenetic networks contribute to many types of cancer (9,10). Lnc00675 is a lncRNA also known as transmembrane protein 238 like (TMEM238L), and is identified as a marker of tumor promoter and unfavorable prognosis in patients with pancreatic ductal adenocarcinoma (11), glioma (12) and cervical cancer (13). In spite of the aforementioned link between Lnc00675 and cancer, very few researches have been carried out to find the molecular mechanism of Lnc00675 in cancer metastasis. Li et al. reported the positively correlation between Lnc00675 expression and TRIP6 protein expression in glioma tissues and cell lines (12). Ma et al. reported that LINC00675 promoted cervical tumorigenesis by modulating the Wnt/β-catenin pathway (13).
However, the association between Lnc00675 and hematological tumors has not been previously reported. In the current study, we analyzed the effect of Lnc00675 on proliferation and apoptosis in human leukemia U937 cells, and the other aim of the current study was to investigate molecular mechanism of Lnc00675 using expression profiling analysis. Our results probably identify Lnc00675 as a novel therapeutic target and provide a new perspective for molecular mechanisms of AML.
Methods
Cell culture and transfection
Human leukemia U937 cells (RRID: CVCL_0007) was cultured in 90% RPMI-1640 (Hyclone, USA) + 10% FBS (Gibco, USA) + penicillin (100 U/mL) and streptomycin (100 g/mL). Cells were cultured under 5% CO2 and 95% air in an incubator set at 37 °C. U937 cells in logarithmic growth phase were divided into three groups, such as Lnc00675 group, si-Lnc00675 group and si-NC group. U937 cells were seeded in 25 cm2 cell culture flasks.
Cell transfection
Transfections were performed using LipofectamineTM 2000 (Invitrogen, USA). U937 cells suspended in serum-free RPMI-1640 were inoculated in 25 cm2 cell culture flasks to undergo transfection with Lnc00675 overexpression vector (Lnc00675 group), Lnc00675 siRNA vector (si-Lnc00675 group), and Lnc00675 siRNA negative control vector (si-NC group), respectively. All nucleotide vectors were purchased from Shanghai Genechem Co., Ltd. (China).
Microarray analysis
U937 cells of Lnc00675 group and si-Lnc00675 were isolated, pelleted cells by centrifugation, respectively. Used 1 mL of TRIzol Reagent (Invitrogen, USA) to lyse 1×107 U937 cells by repetitive pipetting. Microarray experiments were conducted by Shanghai KangChen Biotech (China) with Agilent Human 4x44K Gene Expression Microarray chips with 444,000 probes, the Agilent One-Color Microarray-Based Gene Expression Analysis protocol was used, including total RNA Clean-up and RNA QC, purify the labeled/amplified RNA and labeled cRNA QC, hybridization, microarray Wash, Scanning, extract data using Agilent Feature Extraction software. Bioconductor DESeq2 version 1.12.3 (https://www.rdocumentation.org/packages/DESeq2) was used to identify differentially expressed genes (DEGs) using a fold change (FC) >2 for significant upregulation or significant downregulation and a false discovery rate (FDR) <0.05. A scatter plot was drawn according to the analysis of the DEGs. Gene ontology (GO, www.geneontology.org) analysis was performed to identify the biologic implications of the DEGs. Fisher’s exact test was used to identify the significant GO terms with FDR-adjusted P values. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to identify biologically important pathways associated with the DEGs. Fisher’s exact test was used to select the significant pathways based on P values (P<0.05) and FDR (FDR <0.27).
Cell apoptosis detection by flow cytometry
The cells of Lnc00675 group, si-Lnc00675 group and si-NC group were seeded in 6-well cell culture plate, respectively. After 48 h of transfection, the U937 cells were washed with PBS. Flow cytometry was used to detect the apoptosis rates of the three groups. The staining was performed by Annexin V-FITC/PI double staining method (GenStar, China). Binding buffer was used to resuspend cells, 5 µL of Annexin V-FITC was added, then incubated at room temperature for 15 minutes in the dark. PI staining (5 µL) was added for 5 minutes before detection. A FACS Calibur cell analyzer (BD Biosciences) was used to analyze cell apoptosis and apoptosis rate. The percentages of apoptotic cells including early apoptotic cells (Annexin V+/PI− cells) and late stage apoptotic cells (Annexin V+/PI+ cells) were calculated.
Cell Counting Kit-8 (CCK-8) assay
The viability of U937 cells was detected using CCK-8 assay (Coffit, China). U937 cells (1×105 cells/mL) in the logarithmic growth phase were prepared as cell suspensions using RPMI-1640 containing 10% FBS. Cell suspension (100 µL) was inoculated into a well of 96-well plates. 96-well plate was incubated at 37 °C and 5% CO2 for 24, 48 or 72 h after transfection. CCK-8 solution (10 µL) was added to each well and incubated for 2 h at 37 °C. The absorbance of each well was measured by microplate reader (Shanghai Flash Spectrum Biotechnology, China) at a wavelength of 450 nm. The proliferation rate was calculated using the equation: proliferation rate (%) = (ODtreatment − ODblank)/(ODcontrol − ODblank) ×100%.
Statistical analysis
GO and KEGG analyses were performed using the online database DAVID 6.8 (https://david.ncifcrf.gov/). The difference between 2 groups was determined by unpaired Student’s t‐test using GraphPad prism 8.0 software. The differences were considered statistically significant at P<0.05. All experimental results are presented as the mean ± SD.
Results
Differential gene expression
By comparing Lnc00675 group with si-Lnc00675 group, the microarray analysis determined a total of 1,981 DEGs (FC ≥2) (Figure 1): 866 genes were upregulated and the remaining 1,115 genes were downregulated. Tables 1,2 showed the TOP50 upregulated genes and the TOP50 downregulated genes, respectively.
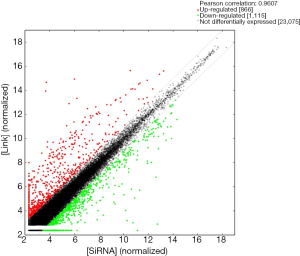
Table 1
| NO. | Gene symbol | Description | Probe Name | GenBank accession | Fold change |
|---|---|---|---|---|---|
| 1 | CCIN | Calicin | A_23_P60227 | NM_005893 | 1,964.76 |
| 2 | LOC100129931 | Uncharacterized LOC100129931 | A_33_P3277883 | NR_033828 | 1,235.66 |
| 3 | CCDC64B | Coiled-coil domain containing 64B | A_33_P3335590 | NM_001103175 | 161.49 |
| 4 | CEP104 | Centrosomal protein 104 kDa | A_33_P3405754 | BC050721 | 59.59 |
| 5 | RAB7A | Member RAS oncogene family | A_33_P3226492 | AF119891 | 51.25 |
| 6 | SFN | Stratifin | A_33_P3389286 | NM_006142 | 51.13 |
| 7 | UTP18 | UTP18 small subunit processome component | A_23_P130020 | NM_016001 | 42.96 |
| 8 | LINC01123 | Long intergenic non-protein coding RNA 1123 | A_33_P3228609 | NR_046110 | 42.82 |
| 9 | LINC01061 | Long intergenic non-protein coding RNA 1061 | A_24_P691775 | NR_037596 | 42.68 |
| 10 | KRTAP1-4 | Keratin associated protein 1-4 | A_33_P3213006 | NM_001257305 | 42.59 |
| 11 | FAM178B | Family with sequence similarity 178 member B | A_33_P3287119 | NM_001122646 | 34.59 |
| 12 | EFTUD1 | Elongation factor Tu GTP binding domain containing 1 | A_24_P754817 | NM_024580 | 33.30 |
| 13 | MAGIX | MAGI family member, X-linked | A_24_P66105 | NM_024859 | 31.73 |
| 14 | DHRS4L1 | Dehydrogenase/reductase SDR family member 4 like 1 | A_33_P3359368 | NM_001277864 | 29.70 |
| 15 | SHISA5 | Shisa family member 5 | A_33_P3270636 | NM_001272068 | 29.61 |
| 16 | SLC51B | Solute carrier family 51, beta subunit | A_23_P436284 | NM_178859 | 28.48 |
| 17 | TBC1D31 | TBC1 domain family, member 31 | A_23_P334218 | NM_145647 | 28.18 |
| 18 | PPP1R14A | Protein phosphatase 1, regulatory (inhibitor) subunit 14A | A_33_P3401647 | NM_033256 | 27.34 |
| 19 | JAKMIP2 | Janus kinase and microtubule interacting protein 2 | A_33_P3255290 | NM_014790 | 26.58 |
| 20 | RAP1GAP2 | RAP1 GTPase activating protein 2 | A_24_P36890 | NM_002885 | 26.24 |
| 21 | OR52E8 | Olfactory receptor, family 52, subfamily E, member 8 | A_33_P3281990 | NM_001005168 | 25.55 |
| 22 | SSPO | SCO-Spondin | A_33_P3277178 | AK093431 | 25.43 |
| 23 | PPP1R1A | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | A_33_P3383471 | AK123969 | 25.30 |
| 24 | MAGEB6 | Melanoma antigen family B, 6 | A_33_P3368755 | NM_173523 | 24.11 |
| 25 | BCR | Breakpoint cluster region | A_24_P127235 | NM_004327 | 24.05 |
| 26 | DCLRE1B | DNA cross-link repair 1B | A_24_P54131 | NM_022836 | 23.81 |
| 27 | RNF150 | Ring finger protein 150 | A_24_P350589 | NM_020724 | 23.74 |
| 28 | HERC6 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 | A_33_P3315779 | NM_001165136 | 23.19 |
| 29 | CUL4A | Cullin 4A | A_33_P3322909 | NM_001278513 | 23.00 |
| 30 | SCOC-AS1 | SCOC antisense RNA 1 | A_24_P145019 | NR_033939 | 22.94 |
| 31 | ATXN3L | Ataxin 3-like | A_23_P361744 | NM_001135995 | 22.73 |
| 32 | BTN3A1 | Butyrophilin, subfamily 3, member A1 | A_33_P3388466 | NM_007048 | 22.47 |
| 33 | AKAP12 | A kinase (PRKA) anchor protein 12 | A_23_P111311 | NM_144497 | 21.77 |
| 34 | CDCA7 | Cell division cycle associated 7 | A_33_P3296169 | NM_031942 | 21.75 |
| 35 | ZDHHC3 | Zinc finger, DHHC-type containing 3 | A_33_P3327479 | NM_016598 | 21.66 |
| 36 | STARD13 | StAR-related lipid transfer (START) domain containing 13 | A_23_P342727 | NM_178006 | 21.51 |
| 37 | CTNND1 | Catenin, delta 1 | A_33_P3209716 | NM_001206885 | 21.20 |
| 38 | PPAN-P2RY11 | PPAN-P2RY11 readthrough | A_33_P3239759 | NM_001198690 | 21.06 |
| 39 | REP15 | RAB15 effector protein | A_33_P3247624 | NM_001029874 | 20.87 |
| 40 | MECP2 | Methyl CpG binding protein 2 | A_33_P3339036 | NM_001110792 | 20.70 |
| 41 | PIK3R5 | Phosphoinositide-3-kinase, regulatory subunit 5 | A_23_P66543 | NM_014308 | 20.65 |
| 42 | THOC2 | THO complex 2 | A_33_P3235690 | NM_001081550 | 20.63 |
| 43 | ZCCHC13 | Zinc finger, CCHC domain containing 13 | A_32_P11096 | NM_203303 | 20.20 |
| 44 | EGFR | Epidermal growth factor receptor | A_33_P3351944 | NM_201283 | 20.01 |
| 45 | FBXO2 | F-box protein 2 | A_23_P45999 | NM_012168 | 19.78 |
| 46 | BICC1 | Bicc family RNA binding protein 1 | A_33_P3293913 | NM_001080512 | 19.61 |
| 47 | PCM1 | Pericentriolar material 1 | A_24_P555510 | NM_006197 | 19.56 |
| 48 | SPECC1L | Sperm antigen with calponin homology and coiled-coil domains 1-like | A_33_P3214027 | NM_001254732 | 19.26 |
| 49 | RIMS3 | Regulating synaptic membrane exocytosis 3 | A_23_P319583 | NM_014747 | 19.16 |
| 50 | TRHDE-AS1 | TRHDE antisense RNA 1 | A_33_P3311493 | NR_026836 | 19.03 |
Table 2
| NO. | Gene symbol | Description | Probe name | GenBank accession | Fold change |
|---|---|---|---|---|---|
| 1 | DBF4B | DBF4 zinc finger B | A_24_P253780 | NM_145663 | 910.03 |
| 2 | KAT2B | K(lysine) acetyltransferase 2B | A_32_P159651 | NM_003884 | 426.94 |
| 3 | FAM50B | Family with sequence similarity 50, member B | A_23_P8240 | NM_012135 | 214.33 |
| 4 | CPSF4L | Cleavage and polyadenylation specific factor 4-like | A_33_P3265194 | – | 135.99 |
| 5 | LOC651337 | Uncharacterized LOC651337 | A_33_P3617190 | AK124119 | 69.68 |
| 6 | NOC3L | Nucleolar complex associated 3 homolog | A_23_P202496 | NM_022451 | 56.46 |
| 7 | MED23 | Mediator complex subunit 23 | A_23_P330999 | NM_015979 | 55.58 |
| 8 | DPH2 | DPH2 homolog | A_24_P393844 | NM_001384 | 47.56 |
| 9 | FOXN2 | Forkhead box N2 | A_32_P140898 | NM_002158 | 28.38 |
| 10 | SLCO3A1 | Solute carrier organic anion transporter family, member 3A1 | A_24_P336276 | NM_013272 | 26.61 |
| 11 | TMEM63A | Transmembrane protein 63A | A_23_P200489 | NM_014698 | 25.86 |
| 12 | RBM5 | RNA binding motif protein 5 | A_23_P18276 | NM_005778 | 25.57 |
| 13 | MAP4 | Microtubule-associated protein 4 | A_23_P211814 | NM_002375 | 22.55 |
| 14 | IRF3 | Interferon regulatory factor 3 | A_23_P27677 | NM_001571 | 18.69 |
| 15 | SUGCT | Succinyl-CoA:glutarate-CoA transferase | A_23_P145711 | NM_024728 | 15.75 |
| 16 | IQSEC2 | IQ motif and Sec7 domain 2 | A_23_P330788 | NM_015075 | 15.01 |
| 17 | ELMOD3 | ELMO/CED-12 domain containing 3 | A_33_P3297302 | NM_001135021 | 14.54 |
| 18 | MEF2BNB | MEF2B neighbor | A_33_P3354771 | AK057161 | 14.32 |
| 19 | UNC80 | Unc-80 homolog | A_33_P3410251 | AK090815 | 14.23 |
| 20 | LRRC8C | Leucine rich repeat containing 8 family, member C | A_33_P3406030 | NM_032270 | 14.13 |
| 21 | ELF4 | E74-like factor 4 | A_24_P340066 | NM_001421 | 13.86 |
| 22 | METTL20 | Methyltransferase like 20 | A_33_P3318966 | NM_173802 | 12.63 |
| 23 | C2CD4C | C2 calcium-dependent domain containing 4C | A_33_P3215412 | NM_001136263 | 12.40 |
| 24 | ZBTB7C | zinc finger and BTB domain containing 7C | A_33_P3402304 | NM_001039360 | 12.04 |
| 25 | NEUROD2 | Neuronal differentiation 2 | A_32_P25295 | NM_006160 | 11.83 |
| 26 | RASD3 | RASD family member 3 | A_33_P3349912 | NM_001257357 | 10.53 |
| 27 | PCDHGC4 | Protocadherin gamma subfamily C, 4 | A_23_P303101 | NM_032406 | 10.50 |
| 28 | TMEM254 | Transmembrane protein 254 | A_23_P97853 | NM_025125 | 10.23 |
| 29 | ANK2 | Ankyrin 2 | A_33_P3287967 | NM_001148 | 9.69 |
| 30 | LOC101928000 | Uncharacterized LOC101928000 | A_33_P3258712 | XR_243583 | 9.43 |
| 31 | SLC31A1 | Solute carrier family 31, member 1 | A_24_P321068 | NM_001859 | 9.39 |
| 32 | LOC100133985 | Uncharacterized LOC100133985 | A_33_P3422654 | NR_024444 | 9.36 |
| 33 | LINC01349 | Long intergenic non-protein coding RNA 1349 | A_33_P3300067 | NR_038914 | 9.27 |
| 34 | CHERP | Calcium homeostasis endoplasmic reticulum protein | A_23_P16139 | NM_006387 | 9.26 |
| 35 | SPRR2C | Small proline-rich protein 2C | A_23_P126089 | NR_003062 | 9.12 |
| 36 | NDRG1 | N-myc downstream regulated 1 | A_23_P20494 | NM_006096 | 9.11 |
| 37 | SLC9A4 | Solute carrier family 9, subfamily A, member 4 | A_33_P3396270 | NM_001011552 | 9.06 |
| 38 | GSTM2P1 | Glutathione S-transferase mu 2 pseudogene 1 | A_23_P58869 | NR_002932 | 8.68 |
| 39 | OR2A2 | Olfactory receptor, family 2, subfamily A, member 2 | A_33_P3394312 | NM_001005480 | 8.68 |
| 40 | PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | A_24_P206604 | NM_004566 | 8.47 |
| 41 | TPSAB1 | Tryptase alpha/beta 1 | A_23_P37702 | NM_003294 | 7.97 |
| 42 | KIF2C | Kinesin family member 2C | A_23_P34788 | NM_006845 | 7.88 |
| 43 | SLC6A8 | Solute carrier family 6, member 8 | A_23_P159937 | NM_005629 | 7.74 |
| 44 | ITGA11 | Integrin, alpha 11 | A_33_P3353791 | NM_181501 | 7.58 |
| 45 | ANXA2R | Annexin A2 receptor | A_33_P3299279 | NM_001014279 | 7.49 |
| 46 | GINS1 | GINS complex subunit 1 | A_33_P3340025 | NM_021067 | 7.49 |
| 47 | LOC283887 | Uncharacterized LOC283887 | A_33_P3677061 | XR_132607 | 7.39 |
| 48 | FAM178A | Family with sequence similarity 178, member A | A_23_P356139 | NM_018121 | 7.24 |
| 49 | CLEC12B | C-type lectin domain family 12, member B | A_33_P3303519 | NM_205852 | 7.11 |
| 50 | GCRG224 | Gastric cancer-related gene GCRG224 | A_33_P3398867 | AF438406 | 7.09 |
GO analysis of the DEGs
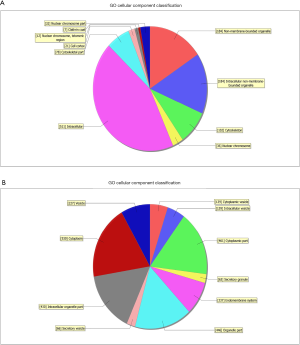
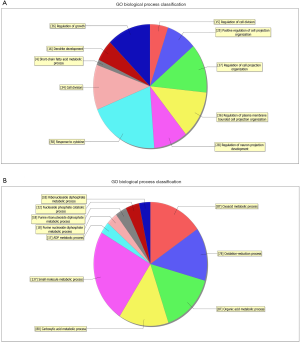
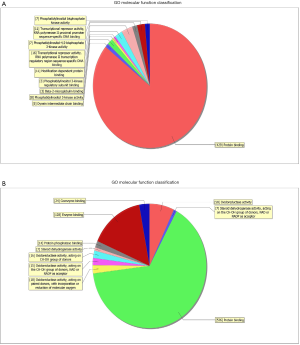
GO analysis contained three domains that represent gene function based on cellular component, biological process and molecular function. A total of 1,385 DEGs were associated with the cell composition domain, of which 608 were upregulated (Figure 2A) and 777 genes were downregulated (Figure 2B). The TOP5 enrichment score biological process terms were “non-membrane-bounded organelle”, “intracellular non-membrane-bounded organelle”, “cytoplasmic vesicle”, “intracellular vesicle” and “cytoplasmic part”. A total of 1,320 DEGs were associated with the biological process domain, of which 581 were upregulated (Figure 3A) and 739 were down-regulated (Figure 3B). The TOP5 enrichment score biological process terms were “oxoacid metabolic process”, “oxidation-reduction process”, “organic acid metabolic process”, “carboxylic acid metabolic process” and “small molecule metabolic process”. A total of 1,324 DEGs were associated with the molecular function domain, of which 580 were upregulated (Figure 4A) and 744 were down-regulated (Figure 4B). The five most enriched molecular function terms were “oxidoreductase activity”, “protein binding”, “steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor”, “protein binding” and “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen”.
Pathway analysis of the DEGs
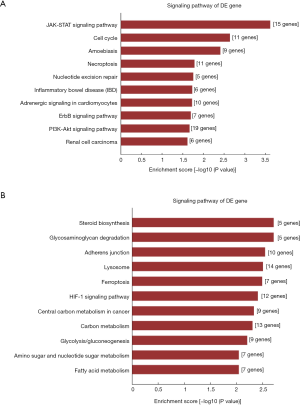
Pathway analysis of the DEGs allows the identification of DEGs related to specific cell pathways. Pathway analysis revealed that DEGs were significantly enriched in 73 pathways (Figure 5A,B). The upregulated genes were involved in 23 pathways and the downregulated DEGs were involved in 50 pathways. The upregulated DEGs were mainly involved in “JAK-STAT signaling pathway”, “Cell cycle”, “Amoebiasis”, “Necroptosis”, “Nucleotide excision repair”, “Inflammatory bowel disease (IBD)”, “Adrenergic signaling in cardiomyocytes”, “ErbB signaling pathway”, “PI3K-Akt signaling pathway” and “Renal cell carcinoma”. The downregulated DEGs were mainly involved in “Steroid biosynthesis”, “Glycosaminoglycan degradation”, “Adherens junction”, “Lysosome, Ferroptosis”, “HIF-1 signaling pathway”, “Central carbon metabolism in cancer”, “Carbon metabolism”, “Glycolysis/Gluconeogenesis”, “Amino sugar and nucleotide sugar metabolism” and “Fatty acid metabolism”.
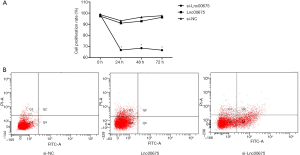
Effects of Lnc00675 on proliferation and apoptosis in U937 cells
The proliferation rate of si-Lnc00675 group was significantly lower than those of si-NC group and Lnc00675 group at all three time points (P<0.05). There was no significant difference in proliferation rate between si-NC group and Lnc00675 group (P>0.05) (Figure 6A). Flow cytometric analysis indicated that the downregulation of Lnc00675 significantly promoted cell apoptosis. The cell apoptosis rate of si-Lnc00675 group (22.93±2.24) was significantly higher than those of si-NC group (0.37±0.88) and Lnc00675 group (0.73±0.35) (P>0.01) (Figure 6B).
Discussion
With the increasing understanding of the lncRNA, the association between tumorigenesis and lncRNA has attracted more and more attention. Notably, Multiple AML researches had shown that the high expression of lncRNA could lead to promote cell proliferation, repress apoptosis, worse prognosis and poor treatment outcomes, such as ZEB2-AS1 (14), lnc-SOX6-1 (15), lnc-CRNDE (16), lnc-HOTAIR (17). With regard to glioma, the high expression of Lnc00675 was dramatically associated with large tumor and advanced World Health Organization grade size (12). The high expression of Lnc00675 positively correlated with poor survival, perineural invasion and lymph node metastasis in patients with pancreatic ductal adenocarcinoma (11). But there is currently no research results available for correlation between Lnc00675 and AML. In the present study, we first reported that the downregulation of Lnc00675 expression resulted in inhibiting cell proliferation and inducing cell apoptosis in U937 cells, but overexpression of Lnc00675 had no effect on the proliferation and apoptosis in U937 cells.
The Wingless (Wnt)/β-catenin signaling pathway has been associated with metabolic reprogramming of cancer cells, cancer stem cells, tumorigenesis and tumor plasticity (18). Ma et al. reported that Lnc00675 inhibited apoptosis and promoted proliferation, migration and invasion though the Wnt/β-catenin pathway in cervical cancer cells, and lithium chloride could attenuate the effects of Lnc00675 knockdown (13). Shan et al. revealed that Lnc00675 downregulated miR-942 expression in colorectal cancer cells, and miR-942 bound to 3’UTR of glycogen synthase kinase-3β (GSK-3β, a kinase mediating β-catenin phosphorylation in Wnt/β-catenin pathway) by dual-luciferase reporter assay (19). At present, there are no studies investigating the molecular mechanism of Lnc00675 in AML cells. In this regard, KEGG pathway analysis were performed using standard enrichment calculation methods to reveal the molecular mechanism. The result of pathway analysis indicated that Lnc00675 involved in JAK-STAT signaling pathway and PI3K-Akt signaling pathway. The activation of JAK-STAT signaling pathway was implicated in the pathogenesis of AML (20,21), and targeting of this pathway was an effective therapeutic strategy for AML (22,23). Dos Santos et al. demonstrated that the PI3K-Akt signaling pathway was constitutively activated in approximately 60% of AML patients cells (24). PI3K-Akt signaling pathway inhibitors, which used alone or with other drugs, have been proven effective for suppressing cell proliferation and promoting apoptosis in AML patients, cell lines or animal models (25).
Epidermal growth factor receptor (EGFR) and interleukin 2 receptor subunit alpha (IL2RA) are involved in both JAK-STAT signaling pathway and PI3K-Akt signaling pathway. Comparing upregulation of Lnc00675 with downregulation of Lnc00675, we found that the expressions of EGFR (FC =20.01) and IL2RA (FC =10.56) were drastically upregulated. EGFR is a cell membrane receptor tyrosine kinase, and mutant EGFR are meaningful serological markers for diagnosis of AML (26). EGFR small molecule inhibitors have been reported to induce complete and durable remission in AML patients (27). Researches indicated a strong association of IL2RA expression with tyrosine kinases pathways. Upregulation of IL2RA expression was correlated with upregulation expressions of fms related receptor tyrosine kinase 3 (FLT3) (28) and inhibitor of DNA binding 1 (ID1) (29), a key target of tyrosine kinases contributing to leukemia transformation. High expression of IL2RA mRNA was an independent and adverse prognostic factor in AML (30).
The present study, to best of our knowledge, was the first to reveal that downregulation of Lnc00675 expression inhibited proliferation and promoted apoptosis in human leukemia U937 cells. By comparing upregulation of Lnc00675 and downregulation of Lnc00675. We identified 866 upregulated DEGs and 1,115 downregulated DEGs, and indicated that Lnc00675 probably affected U937 cells proliferation and apoptosis through JAK-STAT signaling pathway and PI3K-Akt signaling pathway. We will elucidate molecular mechanism of Lnc00675 in AML and further validate the Lnc00675-mediated signaling pathways in our following researches. The results obtained in the current study may aid in the elucidation of molecular mechanisms of Lnc00675 in AML and contribute to the development of target therapies to treat AML.
Acknowledgments
Funding: The present study was supported by grants from
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1714
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1714). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424-47. [Crossref] [PubMed]
- Kadono M, Kanai A, Nagamachi A, et al. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol 2016;44:745-754.e4. [Crossref] [PubMed]
- Mer AS, Lindberg J, Nilsson C, et al. Expression levels of long non-coding RNAs are prognostic for AML outcome. J Hematol Oncol 2018;11:52. [Crossref] [PubMed]
- Morlando M, Fatica A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int J Mol Sci 2018;19:570. [Crossref] [PubMed]
- Tong YS, Zhou XL, Wang XW, et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med 2014;12:233. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol 2017;14:1705-14. [Crossref] [PubMed]
- Li J, Tian H, Yang J, et al. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biol 2016;35:459-70. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018;109:2093-100. [Crossref] [PubMed]
- Li DD, Fu ZQ, Lin Q, et al. Linc00675 is a novel marker of short survival and recurrence in patients with pancreatic ductal adenocarcinoma. World J Gastroenterol 2015;21:9348-57. [Crossref] [PubMed]
- Li Z, Li Y, Wang Q. LINC00675 is a prognostic factor and regulates cell proliferation, migration and invasion in glioma. Biosci Rep 2018;38:BSR20181039. [Crossref] [PubMed]
- Ma S, Deng X, Yang Y, et al. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed Pharmacother 2018;108:1686-93. [Crossref] [PubMed]
- Shi X, Li J, Ma L, et al. Overexpression of ZEB2-AS1 lncRNA is associated with poor clinical outcomes in acute myeloid leukemia. Oncol Lett 2019;17:4935-47. [Crossref] [PubMed]
- Guan X, Wen X, Xiao J, et al. Lnc-SOX6-1 upregulation correlates with poor risk stratification and worse treatment outcomes, and promotes cell proliferation while inhibits apoptosis in pediatric acute myeloid leukemia. Int J Lab Hematol 2019;41:234-41. [Crossref] [PubMed]
- Wang Y, Zhou Q, Ma JJ. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. Eur Rev Med Pharmacol Sci 2018;22:763-70. [PubMed]
- El-Khazragy N, Ghozy S, Matbouly S, et al. Interaction between 12p chromosomal abnormalities and Lnc-HOTAIR mediated pathway in acute myeloid leukemia. J Cell Biochem 2019;120:15288-96. [Crossref] [PubMed]
- El-Sahli S, Xie Y, Wang L, et al. Wnt Signaling in Cancer Metabolism and Immunity. Cancers (Basel) 2019;11:904. [Crossref] [PubMed]
- Shan Z, An N, Qin J, et al. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/β-catenin signaling. Biomed Pharmacother 2018;101:769-76. [Crossref] [PubMed]
- Cook AM, Li L, Ho Y, et al. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood 2014;123:2826-37. [Crossref] [PubMed]
- Gouilleux-Gruart V, Gouilleux F, Desaint C, et al. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood 1996;87:1692-7. [Crossref] [PubMed]
- Faderl S, Ferrajoli A, Harris D, et al. Atiprimod blocks phosphorylation of JAK-STAT and inhibits proliferation of acute myeloid leukemia (AML) cells. Leuk Res 2007;31:91-5. [Crossref] [PubMed]
- Venugopal S, Bar-Natan M, Mascarenhas JO. JAKs to STATs: A tantalizing therapeutic target in acute myeloid leukemia. Blood Rev 2020;40:100634. [Crossref] [PubMed]
- Dos Santos C, Récher C, Demur C, et al. The PI3K/Akt/mTOR pathway: a new therapeutic target in the treatment of acute myeloid leukemia. Bull Cancer 2006;93:445-7. [PubMed]
- Martelli AM, Evangelisti C, Chiarini F, et al. The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget 2010;1:89-103. [Crossref] [PubMed]
- Abdel-Aziz MM. Clinical significance of serum p53 and epidermal growth factor receptor in patients with acute leukemia. Asian Pac J Cancer Prev 2013;14:4295-9. [Crossref] [PubMed]
- Lainey E, Sébert M, Thépot S, et al. Erlotinib antagonizes ABC transporters in acute myeloid leukemia. Cell Cycle 2012;11:4079-92. [Crossref] [PubMed]
- Ozeki K, Kiyoi H, Hirose Y, et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood 2004;103:1901-8. [Crossref] [PubMed]
- Tam WF, Gu TL, Chen J, et al. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood 2008;112:1981-92. [Crossref] [PubMed]
- Du W, He J, Zhou W, et al. High IL2RA mRNA expression is an independent adverse prognostic biomarker in core binding factor and intermediate-risk acute myeloid leukemia. J Transl Med 2019;17:191. [Crossref] [PubMed]


