
Epithelial-mesenchymal transition classification of circulating tumor cells in lung and colon cancer patients: potential role in clinical practice
Introduction
Circulating tumor cells (CTCs) have been shown as a potential biomarker to noninvasively monitor cancer progression and guide therapeutic decisions (1). Primary tumors to release CTCs that travel through the bloodstream and spread to distant sites is responsible for metastasis. Epithelial-mesenchymal transition (EMT) is a key process for cancer metastasis that involves down-regulation of epithelial markers such as epithelial cell adhesion molecule (EpCAM) and cytokeratins (CKs) and up-regulation of mesenchymal markers such as vimentin and TWIST1 in tumor cells. Therefore, the EMT process endows cancer cells with migratory and invasive properties and promotes CTCs formation (2).
CTC isolation and identification techniques are most commonly based on the detection of epithelial markers (i.e., EpCAM and CKs). Epithelial antigen-based approaches could not detect some aggressive CTC subpopulations that have undergone EMT. This has stimulated the detection of EMT-shifted CTCs for monitoring cancer progression and guiding therapy (3). Yu et al. have demonstrated that the increase in mesenchymal CTCs was associated with disease progression, and the shift from mesenchymal phenotype (M phenotype) to epithelial phenotype (E phenotype) was response for therapy in breast cancer (4). Wei et al. have demonstrated that the mesenchymal marker vimentin-positive CTC was an effective biomarker for predicting recurrence-free survival and monitoring the response to treatment in patients with pancreatic cancer (5). Recently, a new CanPatrol CTC-enrichment technique has been reported. This technique can classify CTCs into three subpopulations (E phenotype, M phenotype and hybrid E/M phenotypes) by probing epithelial and mesenchymal markers (e.g., EpCAM, E-cadherin, CK8/18/19, vimentin, and Twist) through RNA in situ hybridization (RNA-ISH) (6). Using the CanPatrol CTC-enrichment technique, Qi et al. have found that CTC count >16 and mesenchymal-CTC percentage >2% in liver cancer were significantly related to the early recurrence, multi-intrahepatic recurrence and lung metastasis (7). These studies suggest that EMT classification of CTC could provide important information for tumor progression.
Since CTCs originate from primary tumors, we firstly investigated the EMT phenotype in lung or colon clinical tumor samples by mining TCGA database. The bioinformatic analysis found that the high expression of mesenchymal markers was associated with poor survival of the patients with lung cancer. However, unlike lung cancer, in colon cancer most of tumor samples exhibited the hybrid expression spectrum of epithelial and mesenchymal markers. Therefore, the new finding based on the bioinformatic analysis promoted us to test whether CTC with mesenchymal phenotype or CTC with hybrid epithelial/mesenchymal phenotypes was an effective biomarker for monitoring tumor progression in lung or colon cancer respectively, by using the Canpatrol CTC EMT analysis technique.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2235).
Methods
Patient samples
From January 2019 to December 2019, a total of 31 patients with lung or colon cancer at the Hefei Cancer Hospital, Chinese Academy of Sciences (Hefei, Anhui, China) were enrolled.
The study was approved by the Ethics Committee of Hefei Cancer Hospital (reference number: 2019KY011) and written informed consent was obtained from all participants. This study was conducted in accordance with the World Medical Association Declaration of Helsinki (as revised in 2013).
CTC test with epithelial and mesenchymal markers
Peripheral blood samples (5 mL) were collected for CTC test using the Canpatrol CTC analysis system (SurExam, Guangzhou, China). Red blood cell lysis buffer was used to remove erythrocytes, and the cells were resuspended in PBS with 4% formaldehyde (Sigma) for 5 minutes. The fixed cells were filtered through a membrane with 8 µm diameter pores (SurExam, Guangzhou, China). The cells on the membrane were subjected to RNA-ISH with the probes, CD45 (leukocyte biomarker), EpCAM and CK8/18/19 (epithelial biomarkers), as well as Vimentin and Twist (mesenchymal biomarkers). DAPI was used to stain the nuclei, and the cells were analyzed with a fluorescent microscope (Zeiss, Germany).
TCGA data mining
By mining the TCGA data of lung adenocarcinoma, lung squamous cell carcinoma and colon adenocarcinoma, mRNA expression levels (z-score threshold ± 2) of epithelial biomarkers (EpCAM and CK8/18/19), and mesenchymal biomarkers (Vimentin and Twist) were analyzed. OncoPrinter and heatmap were generated with the cBioPortal online tool (8,9). The correlation of epithelial and mesenchymal biomarkers’ expression with patients’ survival was generated with the cBioPortal online tool. The expression of epithelial and mesenchymal biomarkers in lung and colon cancers with different stages was generated with GEPIA2 online tool (10).
Statistical analysis
All data were analyzed using PRISM4 Software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis was performed using an unpaired t-test. Results were considered as statistically significant when P<0.05.
Results
TCGA database mining indicates that high expression of mesenchymal markers is associated with poor survival of patients with lung cancer
To indicate the role of EMT in the development of lung cancer, we firstly examined the association of EMT markers with clinicopathological characteristics by mining TCGA database. The OncoPrinter plot indicated that mesenchymal markers (Twist and Vimentin) were highly expressed in 7% and 2.6% of tumor samples of lung adenocarcinoma, as well as in 4% and 4% of tumor samples of lung squamous cell carcinoma. The heatmap showed that epithelial markers (E-cadherin and CK8/18/19) were able to cluster together, while mesenchymal markers (Twist and Vimentin) were clustered together. The tumor samples of lung adenocarcinoma and lung squamous cell carcinoma could be classified into three phenotypes, E phenotype with the expression of epithelial markers, M phenotype with the expression of mesenchymal markers, and hybrid E/M phenotypes with the expression of epithelial and mesenchymal markers (Figure 1A,B).
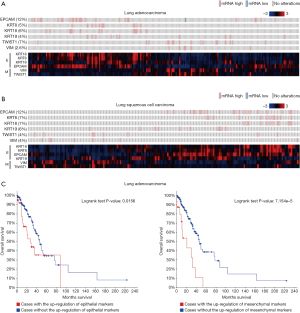
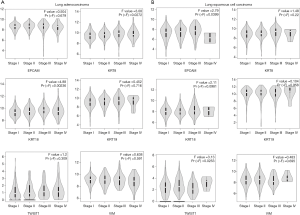
Surprisingly, we found that the high expression of mesenchymal markers in tumor samples was significantly correlated with poor overall survival of patients with lung adenocarcinoma. Although the high expression of epithelial markers was also significantly correlated with poor overall survival, the correlation was more significant in patients with the high expression of mesenchymal markers than patients with the high expression of epithelial markers (Figure 1C). Furthermore, in lung adenocarcinoma, we found that the expression of epithelial markers CK8 and CK18 was significantly higher in tumor samples of advanced stage compared with early stage. In lung squamous cell carcinoma, the expression of mesenchymal marker Twist was significantly higher in tumor samples of stage IV (Figure 2). Taken together, these results suggest that mesenchymal markers are expressed in a subset of lung cancer patients with poor prognosis. Therefore, CTC detection with mesenchymal markers may provide useful information for cancer progression in lung cancer.
TCGA database mining indicates that majority of the tumor samples of colon cancer express hybrid epithelial/mesenchymal markers
We also examined the expression pattern of EMT markers as well as their clinical association in colon cancer by mining TCGA data. The OncoPrinter plot indicated that mesenchymal markers (Twist and Vimentin) were highly expressed in 1.2% and 6% of tumor samples of colon adenocarcinoma. Unlike lung cancer, the heatmap showed that epithelial marker EPCAM was not clustered with other epithelial markers (CK8/18/19). The tumor samples of colon adenocarcinoma could be classified into two clusters, E phenotype with the expression of epithelial markers and hybrid E/M phenotypes with the expression of epithelial and mesenchymal markers. Interestingly, the tumors with hybrid E/M phenotypes can be further classified into two subtypes, hybrid E/M phenotypes with high EPCAM expression as well as hybrid E/M phenotypes with high CK8/CK18/CK19 expressions (Figure 3A).
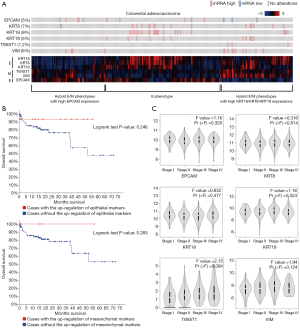
The high expression of epithelial and mesenchymal markers was not correlated with poor overall survival of patients with colon adenocarcinoma (Figure 3B). The expression of epithelial and mesenchymal markers was not correlated with clinical stage in colon cancer (Figure 3C). These results suggest that the expression of mesenchymal markers is coupled with epithelial markers in colon cancer. Therefore, CTCs with hybrid E/M phenotype may provide useful information for monitoring tumor progression in colon cancer.
CTC detection with EMT classification indicates that CTC with mesenchymal phenotype may be predictive biomarkers for tumor progression in lung cancer
Next, we used the Canpatrol CTC analysis system to study whether CTC with mesenchymal phenotype could monitor tumor progression in lung cancer. 16 patients with non-small cell lung cancer with a median age of 60.5 years (range, 40–77 years) were enrolled for the CTC detection. The clinical characteristics and CTC counts (E phenotype, M phenotype and hybrid E/M phenotypes) of the 16 patients were summarized in Tables 1,2. Among the 16 patients, 4 patients (25%) were early stage (I/II), and 12 patients (75%) were advanced stage (III/IV). CTCs were detected in 15/16 patients (94%) and CTC was not detected in one patient with stage II. CTCs with E phenotype, M phenotype and hybrid E/M phenotypes were detected in 8/16 patients (50%), 12/16 patients (75%) and 14/16 patients (87.5%), respectively (Tables 1,2 and Figure 4A). Although the average number of CTC with E phenotype, M phenotype and hybrid E/M phenotypes was higher in advanced stage patients than that in early stage patients, the CTC with M phenotype tended to be statistically significant (Figure 4B). These data indicate that mesenchymal CTC may be a predictive biomarker for advanced lung cancer.
Table 1
| Patient ID | Age | Sex | Stage | Histology | CTC number | |||
|---|---|---|---|---|---|---|---|---|
| Total | E phenotype | M phenotype | Hybrid E/M phenotypes | |||||
| 1# | 62 | M | II | Adenocarcinoma | 3 | 3 | 0 | 0 |
| 2# | 40 | F | I | Adenocarcinoma | 3 | 0 | 1 | 2 |
| 3# | 60 | M | II | Squamous cell carcinoma | 0 | 0 | 0 | 0 |
| 4# | 48 | F | II | Adenocarcinoma | 16 | 0 | 1 | 15 |
| 5# | 55 | F | III | Adenocarcinoma | 5 | 0 | 2 | 3 |
| 6# | 51 | F | IV | Squamous cell carcinoma | 10 | 0 | 8 | 2 |
| 7# | 75 | M | IV | Squamous cell carcinoma | 22 | 5 | 4 | 13 |
| 8# | 61 | F | IV | Adenocarcinoma | 30 | 1 | 6 | 23 |
| 9# | 53 | F | IV | Adenocarcinoma | 19 | 2 | 10 | 7 |
| 10# | 57 | M | IV | Adenocarcinoma | 8 | 0 | 5 | 3 |
| 11# | 58 | F | IV | Adenocarcinoma | 5 | 1 | 3 | 1 |
| 12# | 72 | M | IV | Adenocarcinoma | 22 | 7 | 10 | 5 |
| 13# | 70 | M | IV | Adenocarcinoma | 2 | 1 | 0 | 1 |
| 14# | 74 | M | III | Adenocarcinoma | 9 | 5 | 3 | 1 |
| 15# | 63 | M | IV | Adenocarcinoma | 3 | 0 | 0 | 3 |
| 16# | 74 | M | III | Squamous cell carcinoma | 22 | 0 | 1 | 21 |
CTC, circulating tumor cells; M, male; F, female; E phenotype, epithelial phenotype; M phenotype, mesenchymal phenotype; Hybrid E/M phenotypes, hybrid epithelial/mesenchymal phenotypes.
Table 2
| Patients number | CTC number (Mean) | ||||
|---|---|---|---|---|---|
| Total | E phenotype | M phenotype | Hybrid E/M phenotypes | ||
| Stages | |||||
| I/II | 4 | 6 | 1 | 1 | 4 |
| III/IV | 12 | 13 | 2 | 4 | 7 |
| Histology | |||||
| Adenocarcinoma | 12 | 10 | 2 | 5 | 3 |
| Squamous cell carcinoma | 4 | 13 | 1 | 3 | 9 |
CTC, circulating tumor cells; E phenotype, epithelial phenotype; M phenotype, mesenchymal phenotype; Hybrid E/M phenotypes, hybrid epithelial/mesenchymal phenotypes.
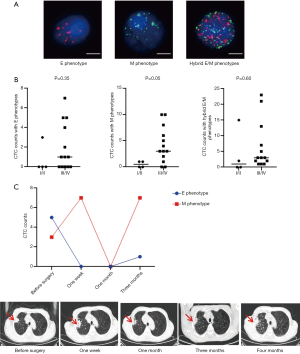
We detected dynamic CTC counts in pre- and post-treatment of a NSCLC patient to monitor the recurrence. A patient (NSCLC, Stage III, 74 years old) was treated with cryo-thermal ablation. After one week of the treatment, the number of CTCs with E phenotype dropped from 5 to 0, and the number of CTCs with M phenotype rose from 3 to 7. One month later, the patient had remarkable clinical response, accompanied by declining CTC (both E and M phenotypes) numbers to 0. After 3 months of this treatment, the patient showed disease progression, which was associated with an increase in both E and M phenotype CTCs (Figure 4C). These data suggest that CTC with M phenotype may effectively predict the cancer progression in lung cancer.
CTC detection with EMT classification indicates that CTC with hybrid E/M phenotypes may be predictive biomarkers for tumor progression in colon cancer
Similarly, we used the Canpatrol CTC analysis system to study whether CTC with hybrid epithelial/mesenchymal phenotypes could monitor tumor progression in colon cancer. 15 patients with colon cancer were enrolled for the CTC detection. The clinical characteristics and CTC counts (E phenotype, M phenotype and hybrid E/M phenotypes) of the 15 patients were summarized in Tables 3,4. Among the 15 patients, 1 patient was adenoma, 2 patients were stage II, 8 patients were stage III and 4 patients were stage IV. CTC were detected in all patients (100%). The positive rate of the E phenotype, M phenotype and hybrid E/M phenotypes was 40.0% (6/15), 66.7% (10/15) and 80.0% (12/15), respectively (Tables 3,4 and Figure 5A). The number of CTCs with E phenotype, M phenotype or hybrid E/M phenotypes was not correlated with the clinical stage in colon cancer (Tables 3,4).
Table 3
| Patient ID | Age | Sex | Histology | Stage | CTC number | |||
|---|---|---|---|---|---|---|---|---|
| Total | E phenotype | M phenotype | Hybrid E/M phenotypes | |||||
| 1# | 67 | M | Adenoma | Benign tumor | 23 | 4 | 9 | 10 |
| 2# | 65 | M | Adenocarcinoma | II | 15 | 0 | 1 | 14 |
| 3# | 51 | M | Adenocarcinoma | II | 31 | 0 | 0 | 31 |
| 4# | 71 | M | Adenocarcinoma | III | 16 | 2 | 6 | 8 |
| 5# | 52 | M | Adenocarcinoma | III | 25 | 0 | 1 | 24 |
| 6# | 39 | M | Mucinous adenocarcinoma | III | 3 | 0 | 3 | 0 |
| 7# | 50 | F | Adenocarcinoma | III | 4 | 0 | 0 | 4 |
| 8# | 46 | M | Mucinous adenocarcinoma | III | 28 | 1 | 4 | 23 |
| 9# | 57 | M | Mucinous adenocarcinoma | III | 44 | 0 | 0 | 44 |
| 10# | 48 | M | Adenocarcinoma | III | 32 | 0 | 3 | 29 |
| 11# | 66 | M | Adenocarcinoma | III | 8 | 1 | 5 | 2 |
| 12# | 69 | F | Adenocarcinoma | IV | 4 | 0 | 1 | 3 |
| 13# | 68 | F | Adenocarcinoma | IV | 20 | 0 | 0 | 20 |
| 14# | 53 | M | Adenocarcinoma | IV | 53 | 1 | 1 | 51 |
| 15# | 60 | M | Adenocarcinoma | IV | 1 | 1 | 0 | 0 |
CTC, circulating tumor cells; M, male; F, female; E phenotype, epithelial phenotype; M phenotype, mesenchymal phenotype; Hybrid E/M phenotypes, hybrid epithelial/mesenchymal phenotypes.
Table 4
| Patients number | CTC number (Mean) | ||||
|---|---|---|---|---|---|
| Total | E phenotype | M phenotype | Hybrid E/M phenotypes | ||
| Stages | |||||
| I/II | 3 | 22 | 1 | 3 | 18 |
| III/IV | 12 | 20 | 1 | 2 | 17 |
| Histology | |||||
| Adenocarcinoma | 12 | 19 | 1 | 2 | 16 |
| mucinous adenocarcinoma | 3 | 25 | 1 | 2 | 22 |
CTC, circulating tumor cells; E phenotype, epithelial phenotype; M phenotype, mesenchymal phenotype; Hybrid E/M phenotypes, hybrid epithelial/mesenchymal phenotypes.
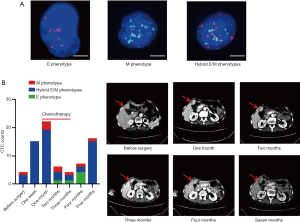
We detected dynamic CTC counts in a patient (colon adenocarcinoma, stage IV, liver and lung metastasis, 69 years old) during her treatment course. The colon tumor of the patient was surgically excised. However, the number of CTCs with hybrid E/M phenotypes increased after one week of surgery. The number of CTCs with hybrid E/M phenotypes still went up after one month of surgery, and then the patient was treated with chemotherapy. After one month of chemotherapy, the patient had remarkable clinical response (liver metastasis shrinked by CT imaging), accompanied by declining CTCs with hybrid E/M phenotypes. However, after two months of completing chemotherapy, the number of CTCs with hybrid E/M phenotypes raised, predicting the likely recurrent. Indeed, the CT imaging indicated that liver metastasis grew bigger after four months of completing chemotherapy (Figure 5B). The data suggest that CTC with hybrid E/M phenotypes may be an effective biomarker for tumor progression in colon cancer.
Discussion
Although many studies have assessed the clinical relevance of the EMT phenotype of CTC subpopulations in different cancers, no study based on bioinformatic analysis of EMT markers combined with CTC detection has been reported so far. In this study, by mining TCGA database, we found that mesenchymal markers were expressed in a subset of clinical tumor samples of lung cancers, and the high expression of mesenchymal markers was associated with poor survival of lung cancer patients. However, in colon cancer, we found that hybrid epithelial/mesenchymal markers were expressed in the majority of clinical tumor samples. Furthermore, we performed the Canpatrol CTC assay to classify EMT phenotypes of CTCs in lung and colon cancer patients. The data of CTC with EMT classification was consistent with the data analyzed by bioinformatics. CTC with M phenotype in lung cancer, and CTC with hybrid E/M phenotypes in colon cancer appeared to be potent for monitoring tumor progression.
The analysis of both bioinformatics and CTC detection demonstrated that mesenchymal phenotype effectively predicted poor clinical characteristics in lung cancer. To our knowledge, this is the first bioinformatic analysis conducted by mining TCGA database, which indicates that high expression of mesenchymal markers is significantly associated with poor survival of lung cancer patients. We used the Canpatrol CTC detection system to further classify EMT phenotypes, and found that in advanced lung cancer patients the number of CTCs with mesenchymal phenotype was higher. Our CTC EMT classification data is consistent with the previous studies. Zhang et al. used the Canpatrol CTC detection system to indicate that CTC with mesenchymal phenotype was the most effective value to differentiate patients with NSCLC with distant metastasis from those with non-distant metastasis (11). Using the CanPatrol CTC EMT classification system, a recent study indicated that lung adenocarcinoma patients with positive mesenchymal CTC had a significantly poor recurrence-free survival and overall survival, compared with the patients with negative mesenchymal CTC (12). Manjunath et al. have demonstrated that CTC expressed higher level of mesenchymal markers compared with matched primary NSCLC tissue, and EMT positive CTC was associated with poorer overall survival of NSCLC patients (13). Morrow et al. have shown that CellSearch EpCAM-dependent platform missed the detection of mesenchymal CTC that contributed the generation of CTC-derived xenografts (14). Therefore, our and previous studies have demonstrated that CTC with mesenchymal phenotype could be a potent biomarker to monitor tumor progression in lung cancer.
Unlike lung cancer, EMT classifications of colon cancer through both TCGA database mining and CTC detection indicated that mesenchymal phenotype was not correlated with advanced stage and poor patients’ survival. A previous study of 126 colon cancer patients using Canpatrol CTC assay has shown that CTCs with mesenchymal phenotype were most common in patients with stage I colon cancer (15). A study including 299 patients with colon cancer has indicated that CTC with positive CK20 detected by RT-PCR was an independent predictor of worse overall survival and disease free survival (16). Furthermore, we found that hybrid epithelial/mesenchymal markers were expressed in the most of tumor samples of colon cancer. The CTC detection with EMT classification indicated that CTC with hybrid E/M phenotype was effective to monitor therapy in a patient with colon cancer. Previous studies have shown that tumor cells with hybrid E/M phenotypes were endowed with stem cell characteristics so that they would be more efficient for metastasis (17). Therefore, our and previous studies have demonstrated that CTC with hybrid E/M phenotypes could be a potent biomarker to monitor tumor progression in colon cancer.
In summary, encouraging results from the current proof-of-concept study indicate that CTCs with M phenotype or hybrid E/M phenotypes could be effective biomarkers for monitoring tumor progression in lung cancer or colon cancer respectively. It is anticipated that a large scale clinical study will be conducted in the future.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2235
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2235
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2235
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2235). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the World Medical Association Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Hefei Cancer Hospital (reference number: 2019KY011) and written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409-24. [Crossref] [PubMed]
- Francart ME, Lambert J, Vanwynsberghe AM, et al. Epithelial-mesenchymal plasticity and circulating tumor cells: Travel companions to metastases. Dev Dyn 2018;247:432-50. [Crossref] [PubMed]
- Gabriel MT, Calleja LR, Chalopin A, et al. Circulating Tumor Cells: A Review of Non-EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin Chem 2016;62:571-81. [Crossref] [PubMed]
- Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4. [Crossref] [PubMed]
- Wei T, Zhang X, Zhang Q, et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett 2019;452:237-43. [Crossref] [PubMed]
- Wu S, Liu S, Liu Z, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One 2015;10:e0123976. [Crossref] [PubMed]
- Qi LN, Xiang BD, Wu FX, et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res 2018;78:4731-44. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Zhang X, Wei L, Li J, et al. Epithelialmesenchymal transition phenotype of circulating tumor cells is associated with distant metastasis in patients with NSCLC. Mol Med Rep 2019;19:601-8. [PubMed]
- Peng H, Tan X, Wang Y, et al. Clinical Significance of Red Cell Distribution Width and Circulating Tumor Cells with an Epithelial-Mesenchymal Transition Phenotype in Lung Adenocarcinoma. Cancer Manag Res 2020;12:5105-17. [Crossref] [PubMed]
- Manjunath Y, Upparahalli SV, Avella DM, et al. PD-L1 Expression with Epithelial Mesenchymal Transition of Circulating Tumor Cells Is Associated with Poor Survival in Curatively Resected Non-Small Cell Lung Cancer. Cancers (Basel) 2019;11:806. [Crossref] [PubMed]
- Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. [Crossref] [PubMed]
- Wu F, Zhu J, Mao Y, et al. Associations between the Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells and the Clinicopathological Features of Patients with Colorectal Cancer. Dis Markers 2017;2017:9474532. [Crossref] [PubMed]
- Hinz S, Hendricks A, Wittig A, et al. Detection of circulating tumor cells with CK20 RT-PCR is an independent negative prognostic marker in colon cancer patients - a prospective study. BMC Cancer 2017;17:53. [Crossref] [PubMed]
- Jolly MK, Boareto M, Huang B, et al. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front Oncol 2015;5:155. [Crossref] [PubMed]


