Screening and verifying key genes with poor prognosis in colon cancer through bioinformatics analysis
Introduction
Colorectal cancer ranks third in incidence and second in mortality across the world (1). There are expected to be 147,950 newly diagnosed colorectal cancer patients in the United States by 2020, including 104,610 cases of colon cancer (CC) and 43,340 cases of rectal cancer. The median age of diagnosis for CC is 69 years, although CC has a trend towards younger age (2). Dietary patterns, body mass index, lifestyle factors will affect the morbidity. The adoption of best practices in cancer treatment and management, longer standing screening, and early detection programs have given rise to the colorectal survival rates in some developed countries. Current screening methods mainly include fecal occult blood test (FOBT), flexible sigmoidoscopy, and colonoscopy. The fecal occult blood test has low specificity. A common colonoscopy is invasive with poor comfort while painless colonoscopy costs more. The preliminary preparation process is more complex. Despite the rapid spread of screening programs since 2000, the proportion of cases diagnosed early in CC has only slightly increased, from 34% in the mid-1990s to 36% between 2012 and 2016 (3). Many researchers, therefore, expect to find CC-specific biomarkers for more convenient and accurate screening. These tumor markers can also be used for cancer risk stratification, treatment protocol formulation, prognostic analysis, and recurrence monitoring.
Understanding the regulation of gene expression is a good way to figure out the relationship between genotype and phenotype (4). Therefore, many scholars applied microarray technology and bioinformatics analysis to medical oncology, hoping to discover the differences between genetic and epigenetic phenotypes caused by tumors and to evaluate markers for disease diagnosis and treatment. However, analyzing independent microarrays from individual surveys often increase the false-positive rate, making it difficult to identify reliable key genes and signaling pathways. Based on this, we expected to screen reliable differentially expressed genes (DEGs) by overlapping related datasets. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2309).
Methods
In this study, we downloaded GSE74602 and GSE10972 gene expression profiles from Gene Expression Omnibus (GEO) (Affymetrix GPL6104 platform, Illumina humanRef-8 v2.0 expression beadchip). There were 30 normal tissues and 30 CC tissues in GSE74602. There were 28 normal tissues and 28 CC tissues in GSE10972. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval from the Institutional Review Board (IRB) was not needed since our data were extracted from the GEO database.
Identification of DEGs
The web tool GEO2R (Http://www.ncbi.nlm.nih.gov/geo/geo2r) was used to screen for DEGs in CC and non-cancer tissue samples. After the standardization of the micro matrix results, DEGs were identified with logFC (fold change) >1 and adj. P value <0.01. A total of 78 up-regulated and 130 down-regulated genes were detected on the ‘Draw Venn Diagram’ website.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses of DEGs
GO and KEGG pathway enrichment analyses were performed by DAVID online database, followed by the construction of the protein-protein interaction (PPI) network. Through the module analysis of the whole network by using the MCODE plug-in of Cytoscape, six hub genes with the highest degree of correlation were screened out.
Survival analysis to screen the candidate genes
Key hub genes and potential pathways were validated in the GEPIA and UALCAN database. This method was helpful to screen out genes with abnormal expression in CC, further understand the initiation and progression of CC at the molecular level, and explore candidate biomarkers for diagnosis, treatment and prognosis.
Statistical analysis
The web tool GEO2R is based on R software version 3.2.3 and the “limma”, “GEOquery” and “Biobase” packages. The GEPIA and UALCAN use the log-rank test, sometimes called the Mantel-Cox test, for the hypothesis evaluation. The GEPIA and UALCAN are based on R (version 3.3.2) and Perl (version 5.22.1) programs (5,6).
Results
Identification of DEGs in CCs
There were 30 normal tissues and 30 CC tissues in GSE74602. There were 28 normal tissues and 28 CC tissues in GSE10972. After standardization of the microarray results, DEGs (1,626 in GSE10972 and 249 in GSE74602) were identified. We used the ‘Draw Venn Diagram’ website to identify the common DEGs in GSE10972 and GSE74602. A total of 78 commonly up-regulated genes (log FC >0) and 130 commonly down-regulated genes were detected (Figure 1A,B & Table 1).
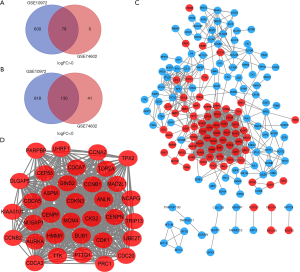
Table 1
| DEGs | Genes name |
|---|---|
| Upregulated | CDH3, SLCO4A1, IQGAP3, TPX2, MMP7, TRIB3, SERPINB5, CCNB1, FOXQ1, MMP3, ASPM, KRT80, LY6G6D, CDCA3, TCN1, ANLN, SLC7A5, CXCL2, CCNB2, PRC1, CDK1, PSAT1, PUS7, NFE2L3, CEP55, CEMIP, TOP2A, CLDN2, VSNL1, PHLDA1, CXCL1, SAPCD2, TMEM97, MCM4, CXCL8, AURKA, SLC12A2, REG1A, MAD2L1, DLGAP5, DPEP1, SOX9, HOXB8, PAICS, HMMR, REG1B, ASCL2, SCD, ETV4, PARPBP, CLDN1, STC2, CDCA7, COL11A1, CDC20, CENPN, CTHRC1UHRF1, ITGA2, KIAA0101, TESC, MYC, CCNA2, TTK, CDKN3, NME1, BUB1, NCAPG, TRIP13, MMP1, PTTG1, CDCA5, GINS2, UBE2T, CENPF, NUSAP1, CKS2, ESM1 |
| Downregulated | PNCK, SPEG, LGALS2, HSPB6, FGL2, OTOP2, DPT, HSPB8, CCL23, MFAP4, ZG16, GUCA2B, WSCD1, CHP2, LRRN2, SCARA5, CLCA4, TCEAL2, ATP1A2, MYOM1, FAM129A, AKR1B10, MT1E, TUBAL3, PRKCB, CA4, CNN1, SEPP1, SDPR, CTSG, TRPM6, NXPE4, ADAMDEC1, ACKR1PY, CCL19, MYLK, HAND1, SLCO2A1, TMIGD1, DNASE1L3, CRYAB, SCIN, CKB, SLC26A3, CD36, CXCL12, PDE5A, MRGPRF, VIP, CA2, FLNC, CHGA, CLEC3B, MT1M, PKIB, ANPEP, ADH1A, CEACAM7, METTL7A, FHL1, ADTRP, PRPH, ST6GALNAC6, MT1G, CCL21, ABCA8, PDE9A, LMO3, TNFRSF13B, ACTG2, MT1F, KRT24, SLC16A9, BEST2, MYL9, INSL5, SLC51B, AQP8, ZBTB16, KCNMB1, DES, FABP4, TNFRSF17, NPTX1, SCGN, CLDN8, MS4A12, FAM107A, PLAC9, MYH11, PGM5, GPX3, SLC25A34, VSIG2, RNF150, NPY, ABI3BP, CALD1, GCNT2, SCNN1B, CASQ2, C2orf88, EPB41L3, CDKN2B, LAMA1, MAMDC2, RBPMS2, BEST4, CA1, SYNM, C7, TEX11, SLC4A4, C2orf40, SFRP1, PDK4, GFRA2, CFD, TMEM37, GPAT3, CPED1, RERGL, CHAIN, SLC51A, GUCA2A, SEMA6A, DHRS9, CA7, AFF3 |
KEGG and GO enrichment analyses of DEGs
We used DAVID software to analyze the biological classification of DEGs. GO-based biological process (BP) analysis indicated that the up-regulated DEGs were particularly enriched in the mitotic nuclear division, cell division, cell proliferation, anaphase-promoting complex-dependent catabolic process and G2/M transition of the mitotic cell cycle, while the down-regulated DEGs were enriched in muscle contraction, bicarbonate transport, cellular response to zinc ion, negative regulation of growth, negative regulation of leukocyte apoptotic process, and one-carbon metabolic process (Table 2). For cell component (CC), the up-regulated DEGs were mainly enriched in the spindle, spindle pole, midbody, nucleus, centrosome, nucleoplasm, and the down-regulated DEGs were significantly enriched in extracellular exosome, extracellular space, Z disc, sarcolemma, extracellular region, extracellular matrix (Table 2). In addition, molecular function (MF) analysis displayed that the up-regulated DEGs were mainly involved in protein kinase binding, chromatin binding, protein binding and cyclin-dependent protein serine/threonine kinase activity, and the down-regulated DEGs were particularly involved in carbonate dehydratase activity, chemokine receptor binding, transporter activity, structural constituent of muscle, chemokine activity and chloride channel activity (Table 2). Furthermore, KEGG analysis results revealed that the up-regulated DEGs were particularly enriched in the cell cycle, oocyte meiosis and progesterone-mediated oocyte maturation, while the down-regulated DEGs were mainly enriched in mineral absorption, bile secretion, nitrogen metabolism, proximal tubule bicarbonate reclamation and pancreatic secretion.
Table 2
| Expression | Category | Term | Count | P value | FDR |
|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_DIRECT | GO:0007067~mitotic nuclear division | 15 | 4.10E−12 | 6.13E−09 |
| GOTERM_BP_DIRECT | GO:0051301~cell division | 16 | 3.32E−11 | 4.96E−08 | |
| GOTERM_BP_DIRECT | GO:0008283~cell proliferation | 11 | 5.04E−06 | 0.007534 | |
| GOTERM_BP_DIRECT | GO:0031145~anaphase-promoting complex-dependent catabolic process | 6 | 2.71E−05 | 0.040467 | |
| GOTERM_BP_DIRECT | GO:0000086~G2/M transition of mitotic cell cycle | 7 | 3.35E−05 | 0.050149 | |
| GOTERM_CC_DIRECT | GO:0005819~spindle | 7 | 1.22E−05 | 0.014011 | |
| GOTERM_CC_DIRECT | GO:0000922~spindle pole | 6 | 9.79E−05 | 0.112351 | |
| GOTERM_CC_DIRECT | GO:0030496~midbody | 6 | 2.16E−04 | 0.247731 | |
| GOTERM_CC_DIRECT | GO:0005634~nucleus | 39 | 2.24E−04 | 0.256455 | |
| GOTERM_CC_DIRECT | GO:0005813~centrosome | 9 | 4.29E−04 | 0.491477 | |
| GOTERM_CC_DIRECT | GO:0005654~nucleoplasm | 24 | 8.62E−04 | 0.985099 | |
| GOTERM_MF_DIRECT | GO:0019901~protein kinase binding | 9 | 2.35E−04 | 0.285395 | |
| GOTERM_MF_DIRECT | GO:0003682~chromatin binding | 8 | 0.001607924 | 1.936332 | |
| GOTERM_MF_DIRECT | GO:0005515~protein binding | 51 | 0.004818525 | 5.700127 | |
| GOTERM_MF_DIRECT | GO:0004693~cyclin-dependent protein serine/threonine kinase activity | 3 | 0.009713212 | 11.1837 | |
| KEGG_PATHWAY | hsa04110: cell cycle | 11 | 5.80E−10 | 6.23E−07 | |
| KEGG_PATHWAY | hsa04114: oocyte meiosis | 8 | 1.63E−06 | 0.001746 | |
| KEGG_PATHWAY | hsa04914: progesterone-mediated oocyte maturation | 6 | 9.13E−05 | 0.09801 | |
| Down-regulated | GOTERM_BP_DIRECT | GO:0006936~muscle contraction | 8 | 6.90E−06 | 0.010446 |
| GOTERM_BP_DIRECT | GO:0015701~bicarbonate transport | 6 | 1.02E−05 | 0.015433 | |
| GOTERM_BP_DIRECT | GO:0071294~cellular response to zinc ion | 4 | 2.52E−04 | 0.381119 | |
| GOTERM_BP_DIRECT | GO:0045926~negative regulation of growth | 4 | 2.52E−04 | 0.381119 | |
| GOTERM_BP_DIRECT | GO:2000107~negative regulation of leukocyte apoptotic process | 3 | 6.38E−04 | 0.962218 | |
| GOTERM_BP_DIRECT | GO:0006730~one-carbon metabolic process | 4 | 0.001002 | 1.506587 | |
| GOTERM_CC_DIRECT | GO:0070062~extracellular exosome | 36 | 3.19E−04 | 0.385946 | |
| GOTERM_CC_DIRECT | GO:0005615~extracellular space | 22 | 3.98E−04 | 0.480914 | |
| GOTERM_CC_DIRECT | GO:0030018~Z disc | 6 | 0.001418 | 1.704013 | |
| GOTERM_CC_DIRECT | GO:0042383~sarcolemma | 5 | 0.002947 | 3.51202 | |
| GOTERM_CC_DIRECT | GO:0005576~extracellular region | 22 | 0.0037 | 4.390367 | |
| GOTERM_CC_DIRECT | GO:0031012~extracellular matrix | 8 | 0.004735 | 5.587538 | |
| GOTERM_MF_DIRECT | GO:0004089~carbonate dehydratase activity | 4 | 1.06E−04 | 0.133058 | |
| GOTERM_MF_DIRECT | GO:0042379~chemokine receptor binding | 3 | 0.001254 | 1.561892 | |
| GOTERM_MF_DIRECT | GO:0005215~transporter activity | 7 | 0.002604 | 3.218176 | |
| GOTERM_MF_DIRECT | GO:0008307~structural constituent of muscle | 4 | 0.002914 | 3.594352 | |
| GOTERM_MF_DIRECT | GO:0008009~chemokine activity | 4 | 0.004518 | 5.52181 | |
| GOTERM_MF_DIRECT | GO:0005254~chloride channel activity | 4 | 0.005935 | 7.194686 | |
| KEGG_PATHWAY | hsa04978: Mineral absorption | 7 | 2.26E−06 | 0.002651 | |
| KEGG_PATHWAY | hsa04976: Bile secretion | 6 | 3.64E−04 | 0.425875 | |
| KEGG_PATHWAY | hsa00910: Nitrogen metabolism | 4 | 4.33E−04 | 0.506185 | |
| KEGG_PATHWAY | hsa04964: Proximal tubule bicarbonate reclamation | 4 | 0.001086 | 1.264063 | |
| KEGG_PATHWAY | hsa04972: Pancreatic secretion | 6 | 0.001429 | 1.659887 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; FDR, false discovery rate.
PPI network and modular analysis
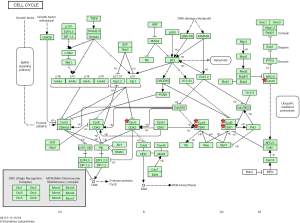
A total of 208 DEGs were included in PPI network analysis. There were 171 nodes and 875 edges mapped in the PPI network for these common DEGs based on STRING database analysis (Figure 1C). The most significant module was obtained by using Cytoscape software. It had 34 nodes and 536 edges (Figure 1D). Genes with degree ≥10 in the module were identified as hub genes. These hub genes were all up-regulated. To further study the potential pathway of DEGs in the module, GO and KEGG enrichment analyses were performed again. The results showed that DEGs in module were mainly enriched in the cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, p53 signaling pathway (Table 3). We choose the top six genes, ranked by degree, including CDK1, CCNB1, CDC20, AURKA, CCNA2, TOP2A. CDK1 had the highest degree with 40, while AURKA, CCNA2 and TOP2A had the lowest degree with 37 (Table 4). Some EDGs (such as CCNB1, CDK1, CDC20, CCNA2, Table 3) were mainly enriched in cell cycle pathway, especially in the G2/M phase (Figure 2).
Table 3
| Category | Term | Count | P value | Genes |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 10 | 2.78E−13 | CCNB1*, CDK1*, MAD2L1, CCNB2, BUB1, TTK, CDC20*, PTTG1, CCNA2*, MCM4 |
| hsa04114 | Oocyte meiosis | 8 | 7.36E−10 | CCNB1*, CDK1*, MAD2L1, CCNB2, BUB1, AURKA*, CDC20*, PTTG1 |
| hsa04914 | Progesterone-mediated oocyte maturation | 6 | 5.28E−07 | CCNB1*, CDK1*, MAD2L1, CCNB2, BUB1, CCNA2* |
| hsa04115 | p53 signaling pathway | 3 | 0.007886 | CCNB1*, CDK1*, CCNB2 |
*, genes were selected hub genes. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 4
| Gene | Degree of connectivity | Regulated |
|---|---|---|
| CDK1 | 40 | Up |
| CCNB1 | 38 | Up |
| CDC20 | 38 | Up |
| AURKA | 37 | Up |
| CCNA2 | 37 | Up |
| TOP2A | 37 | Up |
Survival analysis of selected hub genes
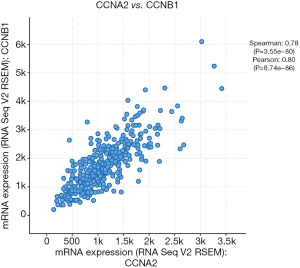
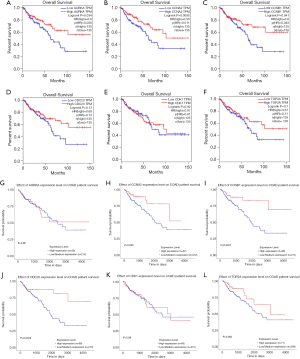
GEPIA (http://gepia.cancer-pku.cn/) was utilized to analyze the survival data of the six hub genes (Figure 3A,B,C,D,E,F). We found that high expression of AURKA, CCNA2 and CCNB1 was significantly associated with worse survival (P<0.05), but CDC20, CDK1 and TOP2A had no significance (P>0.05). And then, UALCAN (http://ualcan.path.uab.edu/) was used to identify six hub genes survival data again (Figure 3G,H,I,J,K,L). The results showed that overexpression of CCNA2, CDC20 and CCNB1 would lead to poor prognosis (P<0.05), while AURKA, CDK1 and TOP2A had no significant correlation with prognosis (P>0.05). Summing up, the data from the two databases (GEPIA and UALCAN) both confirmed that the high expression of CCNA2 and CCNB1 had a significant correlation with poor prognosis, and the expression of these two genes was positively correlated (Figure 4).
Discussion
Despite the significant advances in diagnosis and treatment in the last decade, the prevalence of CC is still rising and five-year survival is still poor (7). Getting more information about the molecular mechanisms of CC is essential for the diagnosis and treatment of CC. DNA microarray technology has improved in sensitivity and selectivity in recent decades and has become a research tool widely used in molecular biology, genome research, cancer diagnosis and other fields (8).
In the present study, we obtained 78 commonly up-regulated genes and 130 commonly down-regulated genes by integrating two expression profiling datasets (GSE10972 and GSE74602) from the GEO dataset. To further understand the role of DEGs play in cancer, we performed the GO and KEGG pathways. The GO term analysis showed that up-regulated DEGs were mainly involved in the regulation of cell division and cell proliferation, while down-regulated DEGs mainly affected the regulation of inflammatory response, cell apoptosis, bicarbonate metabolism and muscle function. The regulation of cell proliferation is crucial to the growth, development and regeneration of eukaryotes, and is considered to be an important cause of cancer (9). Bicarbonate transporters may alter the rate of cancer cell proliferation by controlling the PH of the cytoplasm and extracellular space (10). Festi et al. found that cancer metastasis induced TGF-β release from bone, which in turn caused myasthenia by reducing ca2+ (11). The enriched KEGG pathways of up-regulated DEGs included cell cycle, oocyte meiosis and progesterone-mediated oocyte maturation. The down-regulated DEGs were significantly enriched in mineral absorption, bile and pancreatic secretion, carbon and nitrogen metabolism. Glutamine has multiple roles in cancer as it contributes its carbon backbone to the tricarboxylic acid (TCA) cycle and is an obligatory nitrogen source for purines and pyrimidines (12).
By analyzing the PPI network constructed with DEGs, the hub genes with the highest correlation were screened out, including CDK1, CCNB1, CDC20, AURKA, CCNA2 and TOP2A. Cyclin-dependent kinase 1 (CDK1) is a member of the serine/threonine-protein kinase family that is a vital determinant of mitotic progression. It affects the cell cycle from G2 to M by phosphorylating various substrates (13). CDK1 is overexpressed in a variety of cancers (14,15) and its accumulation in tumors of different sites is positively correlated with the degree of malignancy (16-23). Apart from the function in the cell cycle, CDK1 appears to be involved in protein translation, cell morphogenesis, protein secretion, and nuclear transport (24). CCNB1 works by switching cells from G2 to M but becomes dysfunctional in cancer cells (25). CCNB1 binds to cyclin-dependent kinase 1 (CDK1) to form a complex that phosphorylates the target substrate, and then alters cell cycle progression (26). FOXM1 promotes the development of human liver cancer cells by transcriptional activation of CCNB1, and the high level of CCNB1 is an independent indicator of poor prognosis in hepatocellular carcinoma patients (27). This occurs not only in HCC, but also in breast cancer, head and neck squamous cell carcinoma, and esophageal carcinoma (28). The expression of the CCNB1 gene in invasive pituitary adenoma was significantly higher than that in non-invasive pituitary adenoma, indicating that the CCNB1 gene influences the invasiveness of pituitary adenoma (29). Cell division cycle 20 (CDC20) is a key molecule in the cell cycle as it plays an important role in mitotic exit (30,31). Overexpression of CDC20 may lead to aneuploidy in the tumor cells (32). Abnormal levels of CDC20 expression was observed in varied human cancer, including cutaneous squamous cell carcinoma (33), oral squamous cell carcinoma (34), glioblastoma (35), and gastric cancer, bladder cancer (36), and cervical cancer (37). The overexpression of CDC20 may also indicate a poor prognosis in non-small cell lung cancer (38), pancreatic cancer (39), estrogen receptor-positive breast cancer (40), and hepatocellular carcinoma (41). AURKA encodes a serine-threonine kinase which controls G2/M conversion (42), centrosome maturation (43), and mitotic spindle formation (44). Abundant evidence suggests that inhibition of AURKA expression can effectively suppress cell proliferation, invasion and metastasis in some tumors (45-47). Wen et al. confirmed that the loss of AURKA was sufficient to restrict myelofibrosis and other Primary myelofibrosis phenotypes in vivo (48). Moreover, AURKA is considered to be a valid therapeutic target for neuroblastoma (49) and has been verified in phase I clinical trials (50). CCNA2 accumulates during the G1 phase and functions as a crucial regulatory factor of the G1/S and G2/M transition by activating CDK2 and CDK1 kinases, respectively (51,52). Fischer et al. suggested that p53 down-regulates CCNA2 and other genes through the p53-p21-dreamcde/ChR pathway to block G2/M cell cycle (53). Loukil et al. believed that the dysregulation of CCNA2 and RhoA would lead cells to escape from niche controls resulting in tumor metastasis (54). CCNA2 affects the activity of progesterone receptors through multiple mechanisms making it a key regulator of progesterone-induced breast cancer (55). TOP2A gene is responsible for encoding the ribozyme (type 2 topoisomerase, TOP2A) that plays a key role in the cell cycle. TOP2A can be used as a biomarker to predict the prognosis of a variety of cancers, such as bladder urothelial carcinoma (56), pancreatic cancer (57), prostate cancer (58) and osteosarcomas (59). TOP2A has become a major cellular target for many chemotherapeutic drugs and can be used early to identify patients who may be benefited from adjuvant or neo-adjuvant targeted therapy approaches. Some studies have shown that the change of TOP2A is an independent predictive marker of sensitivity to anthracyclines in adjuvant therapy for early breast cancer (60,61). TOP2A is also used as a marker for the efficacy of pegylated lyposomal doxorubicin (PLD) in epithelial ovarian cancer (62).
In this study, two GSE datasets were selected to screen DEGs and survival analysis was performed in GEPIA and UALCAN databases, which reduced the random errors that may be caused by a single data set and improved the reliability of the results. However, this study also has some limitations. First, the small sample size of this experiment limits the generalization of the conclusion. Second, this result has not been verified in clinical samples due to the accessibility of the data. In the following study, we can collect clinical samples and conduct molecular experiments to verify the six key hub genes we have discovered. In addition, more experiments can be conducted to clarify the upstream regulatory pathways and downstream mechanisms of key differential genes.
In conclusion, there were significant differences in expression levels of CDK1, CCNB1, CDC20, AURKA, CCNA2 and TOP2A between normal and CC tissues. The expression levels of CCNA2 and CCNB1 were negatively correlated with the survival time of patients. CCNA2 and CCNB1 may be potential biomarkers and therapeutic targets for CC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2309
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2309). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval from the Institutional Review Board (IRB) was not needed since our data were extracted from a database.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Su B-B, Shi H, Wan J. Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol 2012;18:2121-6. [Crossref] [PubMed]
- Mantione KJ, Kream RM, Kuzelova H, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med Sci Monit Basic Res 2014;20:138-42. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Li XL, Zhou J, Chen ZR, et al. P53 mutations in colorectal cancer - molecular pathogenesis and pharmacological reactivation. World J Gastroenterol 2015;21:84-93. [Crossref] [PubMed]
- Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng 2002;4:129-53. [Crossref] [PubMed]
- Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, et al. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther 2013;138:255-71. [Crossref] [PubMed]
- Higgins V, Omidi A, Tahmasebi H, et al. Marked Influence of Adiposity on Laboratory Biomarkers in a Healthy Cohort of Children and Adolescents. J Clin Endocrinol Metab 2020;105:e1781-97. [Crossref] [PubMed]
- Festi D, Schiumerini R, Marasco G, et al. Non-invasive diagnostic approach to non-alcoholic fatty liver disease: current evidence and future perspectives. Expert Rev Gastroenterol Hepatol 2015;9:1039-53. [Crossref] [PubMed]
- Bott AJ, Maimouni S, Zong WX. The Pleiotropic Effects of Glutamine Metabolism in Cancer. Cancers (Basel) 2019;11:770. [Crossref] [PubMed]
- Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015;14:130-46. [Crossref] [PubMed]
- Dai W, Li Y, Meng X, et al. Does tumor size have its prognostic role in colorectal cancer? Re-evaluating its value in colorectal adenocarcinoma with different macroscopic growth pattern. Int J Surg 2017;45:105-12. [Crossref] [PubMed]
- Yokoyama M, Watanabe T, Otaki Y, et al. Association of the Aspartate Aminotransferase to Alanine Aminotransferase Ratio with BNP Level and Cardiovascular Mortality in the General Population: The Yamagata Study 10-Year Follow-Up. Dis Markers 2016;2016:4857917. [Crossref] [PubMed]
- Corrêa MM, Thumé E, De Oliveira ERA, et al. Performance of the waist-to-height ratio in identifying obesity and predicting non-communicable diseases in the elderly population: A systematic literature review. Arch Gerontol Geriatr 2016;65:174-82. [Crossref] [PubMed]
- Denney-Wilson E, Hardy LL, Dobbins T, et al. Body mass index, waist circumference, and chronic disease risk factors in Australian adolescents. Arch Pediatr Adolesc Med 2008;162:566-73. [Crossref] [PubMed]
- Hongo F, Takaha N, Oishi M, et al. CDK1 and CDK2 activity is a strong predictor of renal cell carcinoma recurrence. Urol Oncol 2014;32:1240-6. [Crossref] [PubMed]
- Lee DE, Lee KW, Jung SK, et al. 6,7,4’-trihydroxyisoflavone inhibits HCT-116 human colon cancer cell proliferation by targeting CDK1 and CDK2. Carcinogenesis 2011;32:629-35. [Crossref] [PubMed]
- Pavlou MP, Dimitromanolakis A, Martinez-Morillo E, et al. Integrating meta-analysis of microarray data and targeted proteomics for biomarker identification: application in breast cancer. J Proteome Res 2014;13:2897-909. [Crossref] [PubMed]
- Willder JM, Heng SJ, McCall P, et al. Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br J Cancer 2013;108:139-48. [Crossref] [PubMed]
- Yang W, Cho H, Shin HY, et al. Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget 2016;7:49481-97. [Crossref] [PubMed]
- Shi YX, Zhu T, Zou T, et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget 2016;7:85235-43. [Crossref] [PubMed]
- Holt LJ, Tuch BB, Villen J, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009;325:1682-6. [Crossref] [PubMed]
- Park TJ, Kim JY, Oh SP, et al. TIS21 negatively regulates hepatocarcinogenesis by disruption of cyclin B1–Forkhead box M1 regulation loop. Hepatology 2008;47:1533-43. [Crossref] [PubMed]
- Fang L, Du WW, Awan FM, et al. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett 2019;459:216-26. [Crossref] [PubMed]
- Chai N, Xie HH, Yin JP, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun 2018;500:924-9. [Crossref] [PubMed]
- Yang Q, Manicone A, Coursen JD, et al. Identification of a functional domain in a GADD45-mediated G2/M checkpoint. J Biol Chem 2000;275:36892-8. [Crossref] [PubMed]
- Li B, Cheng J, Wang H, et al. CCNB1 affects cavernous sinus invasion in pituitary adenomas through the epithelial-mesenchymal transition. J Transl Med 2019;17:336. [Crossref] [PubMed]
- Chang L, Zhang Z, Yang J, et al. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 2015;522:450-4. [Crossref] [PubMed]
- Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. J Cell Biol 2013;201:177-89. [Crossref] [PubMed]
- Mondal G, Sengupta S, Panda CK, et al. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis 2007;28:81-92. [Crossref] [PubMed]
- Chu Z, Zhang X, Li Q, et al. CDC20 contributes to the development of human cutaneous squamous cell carcinoma through the Wnt/betacatenin signaling pathway. Int J Oncol 2019;54:1534-44. [PubMed]
- Moura IM, Delgado ML, Silva PM, et al. High CDC20 expression is associated with poor prognosis in oral squamous cell carcinoma. J Oral Pathol Med 2014;43:225-31. [Crossref] [PubMed]
- Mao DD, Gujar AD, Mahlokozera T, et al. A CDC20-APC/SOX2 Signaling Axis Regulates Human Glioblastoma Stem-like Cells. Cell Rep 2015;11:1809-21. [Crossref] [PubMed]
- Choi JW, Kim Y, Lee JH, et al. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch 2013;463:681-7. [Crossref] [PubMed]
- Kim Y, Choi JW, Lee JH, et al. MAD2 and CDC20 are upregulated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the uterine cervix. Int J Gynecol Pathol 2014;33:517-23. [Crossref] [PubMed]
- Kato T, Daigo Y, Aragaki M, et al. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol 2012;106:423-30. [Crossref] [PubMed]
- Collaboration NCDRF. Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016;387:1513-30. [Crossref] [PubMed]
- Alfarsi LH, Ansari RE, Craze ML, et al. CDC20 expression in oestrogen receptor positive breast cancer predicts poor prognosis and lack of response to endocrine therapy. Breast Cancer Res Treat 2019;178:535-44. [Crossref] [PubMed]
- Li J, Gao JZ, Du JL, et al. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int J Oncol 2014;45:1547-55. [Crossref] [PubMed]
- Kaestner P, Stolz A, Bastians H. Determinants for the efficiency of anticancer drugs targeting either Aurora-A or Aurora-B kinases in human colon carcinoma cells. Mol Cancer Ther 2009;8:2046-56. [Crossref] [PubMed]
- Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer 2005;5:42-50. [Crossref] [PubMed]
- Hirota T. Aurora-A and an interacting Activator, the LIM Protein Ajuba, Are Required for Mitotic Commitment in Human Cellls. Cell 2003;114:585-98. [Crossref] [PubMed]
- Li Y, Zhang ZF, Chen J, et al. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma Am J Transl Res 2010;2:296-308. [PubMed]
- Wang X, Dong L, Xie J, et al. Stable knockdown of Aurora-A by vector-based RNA interference in human esophageal squamous cell carcinoma cell line inhibits tumor cell proliferation, invasion and enhances apoptosis. Cancer Biol Ther 2009;8:1852-9. [Crossref] [PubMed]
- Addepalli MK, Ray KB, Kumar B, et al. RNAi-mediated knockdown of AURKB and EGFR shows enhanced therapeutic efficacy in prostate tumor regression. Gene Ther 2010;17:352-9. [Crossref] [PubMed]
- Wen QJ, Yang Q, Goldenson B, et al. Targeting megakaryocytic-induced fibrosis in myeloproliferative neoplasms by AURKA inhibition. Nat Med 2015;21:1473-80. [Crossref] [PubMed]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010;362:2202-11. [Crossref] [PubMed]
- Mossé YP, Lipsitz E, Fox E, et al. Pediatric phase I trial and pharmacokinetic study of MLN8237, an investigational oral selective small-molecule inhibitor of Aurora kinase A: a Children’s Oncology Group Phase I Consortium study. Clin Cancer Res 2012;18:6058-64. [Crossref] [PubMed]
- Krasnov GS, Puzanov GA, Kudryavtseva AV, et al. Differential Expression of an Ensemble of the Key Genes Involved in Cell-Cycle Regulation in Lung Cancer. Mol Biol (Mosk) 2017;51:849-56. [PubMed]
- Pagano M, Pepperkok R, Verde F, et al. Cyclin A is required at two points in the human cell cycle. EMBO J 1992;11:961-71. [Crossref] [PubMed]
- Fischer M, Quaas M, Steiner L, et al. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res 2016;44:164-74. [Crossref] [PubMed]
- Loukil A, Cheung CT, Bendris N, et al. Cyclin A2: At the crossroads of cell cycle and cell invasion. World J Biol Chem 2015;6:346-50. [Crossref] [PubMed]
- Moore NL, Edwards DP, Weigel NL. Cyclin A2 and its associated kinase activity are required for optimal induction of progesterone receptor target genes in breast cancer cells. J Steroid Biochem Mol Biol 2014;144 Pt B:471-82.
- Zeng S, Liu A, Dai L, et al. Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer 2019;19:604. [Crossref] [PubMed]
- Pei YF, Yin XM, Liu XQ. TOP2A induces malignant character of pancreatic cancer through activating beta-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis 2018;1864:197-207. [Crossref] [PubMed]
- Labbé DP, Sweeney CJ, Brown M, et al. TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup. Clin Cancer Res 2017;23:7072-83. [Crossref] [PubMed]
- Nguyen A, Lasthaus C, Guerin E, et al. Role of Topoisomerases in Pediatric High Grade Osteosarcomas: TOP2A Gene Is One of the Unique Molecular Biomarkers of Chemoresponse. Cancers (Basel) 2013;5:662-75. [Crossref] [PubMed]
- Bartlett JM, McConkey CC, Munro AF, et al. Predicting Anthracycline Benefit: TOP2A and CEP17-Not Only but Also. J Clin Oncol 2015;33:1680-7. [Crossref] [PubMed]
- Di Leo A, Desmedt C, Bartlett JM, et al. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol 2011;12:1134-42. [Crossref] [PubMed]
- Ghisoni E, Maggiorotto F, Borella F, et al. TOP2A as marker of response to pegylated lyposomal doxorubicin (PLD) in epithelial ovarian cancers. J Ovarian Res 2019;12:17. [Crossref] [PubMed]