
TM9SF4 expression in tumor tissues: a novel diagnostic biomarker for gastrointestinal tumors
Introduction
Gastric cancer (GC) and colorectal cancer (CRC) are respectively the fifth and the third most common malignancies globally, as well as the third and second leading cause of cancer related deaths (1). Survival of patients affected by these cancers is significantly associated with disease staging at diagnosis. The five-year survival rate for CRC patients diagnosed in early stage is >90%, while for those diagnosed in late stage is approximately 7%. Currently, there are several approaches to screen these cancers, mainly based on endoscopic analysis of the mucosae followed by biopsy and fecal occult blood test (FOBT) for colon cancer (2-4). These techniques have inherent limitations, especially in early diagnosis settings: endoscopic exams are invasive, costly and associated with patients discomfort and procedural risks, while FOBT, though non-invasive and affordable, has inadequate sensitivity and specificity to stand alone as a diagnostic test (5,6). For these reasons, a considerable amount of these tumors is diagnosed at advanced stages when the complete cure is hardly possible. Diagnostic delay is also due to a lack of symptoms and of specific biomarkers. In facts, analysis of tumor biopsies for suspected GC and CRC is currently based on pathologist examination of counterstained tissue, with no immunohistochemistry (IHC) markers being widely recognized as useful for early diagnostics (7). Development of novel sensitive biomarkers for assessing occult residual diseases after surgery and monitoring recurrences or metastases of CRC is therefore urgently needed.
Carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9) are among the best available prognostic markers for digestive cancers, including GC and CRC (8-10). However, low sensitivity and low specificity of these markers affects their use as biomarkers for early diagnosis, and their expression level is considered for post-resection monitoring of patients with already diagnosed cancer.
Transmembrane 9 Superfamily 4 (TM9SF4) is a recently described transmembrane protein highly expressed in melanoma, colon carcinoma cell lines, acute myeloid leukemia and myelodysplastic syndromes. Recent studies have demonstrated that TM9SF4 is an autophagic marker involved in tumor cells cannibalism, a phenomenon often related to poor prognosis (11-15). TM9SF4 is mainly detectable in intracellular vesicles such as endosomes and other vesicles, where aberrantly activates the H[+]-Vacuolar ATPase, a proton pump involved in the tumor pH gradient alterations associated with drug resistance and invasiveness of cancer cells (16).
The aim of our study was: (I) to analyze the TM9SF4 expression in GC, CRC tissues and precancerous lesions associated to these cancers, (II) evaluate its diagnostic value in comparison to CEA and CA 19-9. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-516).
Methods
Patients
One hundred and eight cancer patients (68 with GC and 40 with CRC) and a relevant group of controls were included to the study. The GC patient cohort had equal proportions of intestinal and diffuse cancers, further subdivided into non-advanced (Stage I–II) and advanced (Stage III–IV) cases (17 cases for each subgroup). Only tissue samples obtained by surgical resection of the tumor were used in this cohort. The control group, defined according to the updated Sydney classification (17), contained 10 patients from whom upper endoscopy biopsy material was available. In addition, 10 patients with low grade dysplasia were enrolled.
Forty surgery samples from patients with CRC were included to the analysis, out of these 20 were with non- advanced (Stage I–II) and 20 with advanced (Stage III–IV) disease. All patients were staged according to the American Joint Committee on Cancer (18). The control group contained 10 patients for whom colonoscopy data were available. Furthermore, 10 patients with hyperplastic and sessile serrated polyp and 10 patients with tubular/villous adenoma were enrolled in the study. All patients signed an informed consent before being included to the study. The study was approved by a local ethical committee (the Central Medical Ethics Committee of Latvia, Approval No. 01-29.1/20 from 22.09.2011) in accordance to the Declaration of Helsinki (as revised in 2013).
The mean age of patients with GC was 61.91±11.38 years, while the male/female ratio was 32/35. The mean age of patients with control group and low grade dysplasia was respectively, 56.45±16.30 and 59.34±12.48 years. Table S1 shows an overview of enrolled patients’ characteristics. The mean age of CRC patients was 64.25±9.565 years, while the male/female ratio was 21/19. The mean age of patients with control group, hyperplastic polyps, sessile serrated adenoma and tubular/villous adenoma was respectively 52.40±18.10, 58.30±14.68, 52.18±18.20 and 61.30±14.49 years. Table S1 shows an overview of enrolled patients characteristics.
Tissue processing and IHC
Paraffin embedded tissue specimens were retrieved from the Biobank of Riga East University Hospital. Specimens were cut in 3-µm-thick sections and slides were stained with haematoxylin and eosin to evaluate histopathological changes. Antigen retrieval was achieved by incubating the slides with Tris/EDTA buffer at pH=9.0 for 30 min in a scientific microwave. The slides were then incubated overnight at 4 °C with rabbit polyclonal anti-TM9SF4 antibody (HansaBioMed OU, Tallinn Estonia), anti-CEA antibody (clone II -7, M7072. DAKO Glostrup Denmark,) and anti-CA 19-9 antibody (clone 116-NS-19-9, M3517, DAKO). Antibody binding was detected using the EnVision reagent following the manufacturer’s instructions (DAKO).
Imaging, quantitation of cells and data analysis
Immunostained slides of each histology sample were scanned with Panoramic Midi slide scanner (3D Histech Hungary) at magnification ×20. The whole-area scanned of each slide was analyzed with Image Analysis QuantCenter (3DHistech). A score was given for every specimen according to the intensity of the nuclear or cytoplasmic staining (no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3) and the extent of stained cells (0% = score 0; 1–10% = 1; 11–50% = 2; >51% = score 3. Negative means 0% area staining. Focally positive means 1–80% area staining, diffusely positive means 81–100% area staining. Values were expressed as mean ± SD.
Statistical analysis
Immunohistochemical data were analyzed by two-way ANOVA followed by Bonferroni post test for comparison between the groups. SPSS 21 version software was used for the statistical analysis.
Results
Tissue specimens from 68 GC and 40 CRC cancer patients and from relevant control groups were used to assess TM9SF4 expression by IHC (19) (Table S1 for patient’s characteristics). The results revealed that, while mild focal staining of TM9SF4 was observed in healthy gastric tissues with an average score of 1.1+0.74 (Figure 1A), TM9SF4 expression was significantly increased in both diffuse and intestinal GC types (Figure 1 B,C). Overall, 90% of all GC samples were found positive with an average score close to 3 (Figure 1D). Importantly, more than 70% of the intestinal type and 60% of diffuse type GC were found to be highly positive for TM9SF4 expression (with a score value of 3+, Figure 1D). Statistical analysis on TM9SF4 staining intensity scores, clearly showed a significant increase of TM9SF4 expression in both early and late GC (P<0.001, two way ANOVA followed by Bonferroni post test; Figure 1D and Figure S1A, left column).
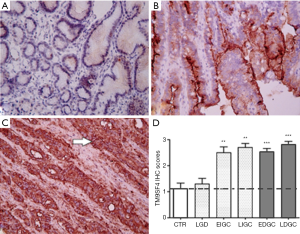
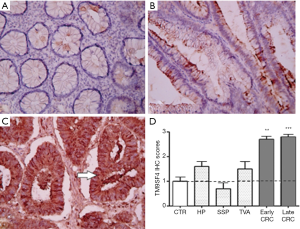
Similarly to gastric tissues, a mild focal staining was observed in the colon mucosa samples of the control group (Figure 2A), while an increase of TM9SF4 expression was observed in both early and late stage CRC (Figure 2B,C). Positive TM9SF4 staining was observed in 85% of CRC tissue samples, with over 80% of them being found highly positive (Score 3+) (Figure 2D and Figure S1A, right column).
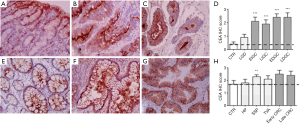
The expression of TM9SF4 was compared to CEA and CA 19-9 in the same biopsies. With regard to CEA, while poorly detectable in gastric mucosa of the control group (Figure 3A), its expression increased in low grade dysplasia and was significantly higher in both early and advanced intestinal and diffuse type GC (Figure 3B,C respectively). Overall, 90% of gastric carcinomas were CEA positive, with IHC scores between 2 and 3 (Figure 3D and Figure S1B left). In colon, CEA was mildly expressed in control samples (colon mucosa; Figure 3E) and, while increased, the levels of this biomarker in both early and advanced CRC did not significantly differ from the control (Figure 3F,G,H). No differences of expression were observed between control and hyperplastic polyps, or tubular villous adenomas while CEA expression was found significantly up-regulated in sessile serrated polyps (Figure 3H and Figure S1B right).
As far as CA 19-9 is concerned, absent or mild focal staining was observed in the gastric mucosa of the control group and in low grade dysplasia samples (Figure 4A and B, respectively), while its expression was found significantly higher in advanced intestinal GC and diffuse GC regardless disease stage (Figure 4C and D). However, only 50% of GC samples expressed CA 19-9 after antibody staining, with a mean score between 1 and 2 (Figure 4D and Figure S1C left). In colon samples (Figure 4E,F,G,H), CA 19-9 expression was increased in 70% of tubular villous adenoma and early stage colorectal carcinoma samples (Figure 4F and G, respectively) with a mean score of about 2 but these differences did not reach statistical significance (Figure 4H and Figure S1C right). Hyperplastic polyps and sessile serrated polyps and late stage CRC were also found weakly positive (Figure 4H) while the control group was completely unstained, indicating no CA 19-9 expression (Figure 4E).
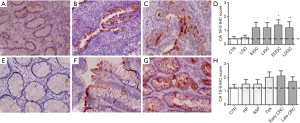
In colon samples, CA 19-9 expression was increased in 70% of tubular villous adenoma and early stage colorectal carcinoma samples (Figure 4F and G, respectively) with a mean score of about 2 but these differences did not reach statistical significance (Figure 4H and Figure S1C right). Hyperplastic polyps and sessile serrated polyps and late stage CRC were also found weakly positive (Figure 4H) while the control group was completely unstained, indicating no CA 19-9 expression (Figure 4E).
In summary, this study shows that TM9SF4 is significantly over-expressed in both non advanced and advanced GC specimens and its expression levels are comparable to CEA and higher than CA 19-9 in these diseased tissues. Furthermore, TM9SF4 is the only biomarker to be upregulated in non-advanced cancer samples, providing a significant advantage over the other two clinically adopted biomarkers.
Conclusions
Notwithstanding the discovery of several molecules as candidate biomarkers for GC and CRC, their adoption in the clinical setting has been limited due to their poor performances in early detection and screening (20,21).
Limited evidence is so far available with respect to the TM9SF4 expression in cancer cells (11-13,22). Recently TM9SF4 has been identified as structurally linked to V-ATPase assembly and activation (11) mediating thus tumor pH alterations that are typical of all malignant cancer cells, and are integral part of a general metabolic switch described under the term of “Warburg Effect” (23).
In the present study, we investigated the expression of TM9SF4 and its potential as a novel tumor marker in gastric and colon cancers. To the best of our knowledge, this is the first study that describes the expression of TM9SF4 in normal gastric and colon tissue, as well as in premalignant lesions and GC and CRC. IHC assessment of TM9SF4 in subjects with CRC and GC compared to healthy surrounding tissue and pre-neoplastic lesions revealed highly specific staining of tumor tissue in both advanced and non-advanced disease, with none or little expression in healthy or dysplastic tissues. Compared with CEA and CA 19-9 markers, TM9SF4 shows higher sensitivity and specificity to identify GC and CRC cases in IHC.
While all three markers are significantly upregulated in both cancer types with respect to healthy mucosa, TM9SF4 was the only one to be significantly upregulated, with a mean score of nearly 3+, in 90% of both cancer types, and the level of expression significantly correlated with disease stage, while absent or poorly detectable in dysplastic control tissues. Since the promoter of TM9SF4 gene contains one highly conserved hypoxic-responsive element it is tempting to speculate that hypoxia of cancer tissue may, at least in part, modulate TM9SF4 expression during disease progression (13).
Correlation between TM9SF4 expression and disease progression is consistent with recent reports suggesting the interaction of TM9SF4 with the V-ATPase, a proton pump highly expressed in GC and CRC cancer (11,24); the TM9SF4/V-ATPase association aberrantly activates V-ATPase that plays a key role in tumor invasion and metastasis by activating several types of proteases including cathepsins, metalloproteases, and gelatinases that degenerate extracellular matrix (16,25).
In conclusion, our data strongly support the potential of TM9SF4 as a new tumor marker for the early diagnosis of GIC, providing the basis for improved screening and classification approaches. Further studies on extended patient cohorts, additional tumor types and sample types (e.g., tissue, plasma serum) are warranted to fully assess the potential of this protein as cancer marker for early diagnostic and prognostic purposes.
Acknowledgments
Funding:
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-516
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-516
Conflicts of interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-516). Dr. Guazzi reports he is an employee of HansaBioMed OU, a company that sells TM9SF4 antibodies. Dr. Chiesi and Dr. Zarovni report that they are inventors of the patent “Monoclonal antibodies, hybridomas nd methods for use”. Pubbl n. 20120122118 licensed to Exosomics spa and Dr Antonio Chiesi is an employee of HansaBioMed OU (HBM) a company that sells TM9SF4 antibodies. Dr. Lozupone reports he is one inventor of the patent “Monoclonal antibodies, hybridomas nd methods for use”. Pubbl n. 20120122118 licensed to Exosomics Spa. The other authors have nothing to disclose.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Central Medical Ethics Committee of Latvia, Approval No. 01-29.1/20 from 22.09.2011. All patients signed an informed consent before being included to the study. Tissue samples have been collected from the Biobank of Riga East University Hospital & Institute of Clinical and Preventive Medicine, University of Latvia.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Leja M, You W, Camargo MC, et al. Implementation of gastric cancer screening - the global experience. Best Pract Res Clin Gastroenterol 2014;28:1093-106. [Crossref] [PubMed]
- Leung WK, Wu MS, Kakugawa Y, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol 2008;9:279-87. [Crossref] [PubMed]
- Vega P, Valentin F, Cubiella J. Colorectal cancer diagnosis: Pitfalls and opportunities. World J Gastrointest Oncol 2015;7:422-33. [Crossref] [PubMed]
- Dickinson BT, Kisiel J, Ahlquist DA, et al. Molecular markers for colorectal cancer screening. Gut 2015;64:1485-94. [Crossref] [PubMed]
- Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol 2014;20:13842-62. [Crossref] [PubMed]
- Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 2006;24:5313-27. [Crossref] [PubMed]
- Duffy MJ, Lamerz R, Haglund C, et al. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer 2014;134:2513-22. [Crossref] [PubMed]
- Wang T, Matsuda Y, Seki A, et al. Carbohydrate Antigen 19-9-Positive Gastric Adenocarcinoma: Autopsy Findings and Review of the Literature. Case Rep Gastroenterol 2017;11:545-53. [Crossref] [PubMed]
- Wu KL, Huang EY, Yeh WL, et al. Synergistic interaction between galectin-3 and carcinoembryonic antigen promotes colorectal cancer metastasis. Oncotarget 2017;8:61935-43. [Crossref] [PubMed]
- Lozupone F, Borghi M, Marzoli F, et al. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene 2015;34:5163-74. [Crossref] [PubMed]
- Lozupone F, Perdicchio M, Brambilla D, et al. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep 2009;10:1348-54. [Crossref] [PubMed]
- Paolillo R, Spinello I, Quaranta MT, et al. Human TM9SF4 Is a New Gene Down-Regulated by Hypoxia and Involved in Cell Adhesion of Leukemic Cells. PLoS One 2015;10:e0126968. [Crossref] [PubMed]
- Perrin J, Mortier M, Jacomin AC, et al. The nonaspanins TM9SF2 and TM9SF4 regulate the plasma membrane localization and signalling activity of the peptidoglycan recognition protein PGRP-LC in Drosophila. J Innate Immun 2015;7:37-46. [Crossref] [PubMed]
- Sun L, Meng Z, Zhu Y, et al. TM9SF4 is a novel factor promoting autophagic flux under amino acid starvation. Cell Death Differ 2018;25:368-79. [Crossref] [PubMed]
- Cotter K, Stransky L, McGuire C, et al. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem Sci 2015;40:611-22. [Crossref] [PubMed]
- Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 1996;20:1161-81. [Crossref] [PubMed]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4.
- Wong HH, Chu P. Immunohistochemical features of the gastrointestinal tract tumors. J Gastrointest Oncol 2012;3:262-84. [PubMed]
- McLaughlin R, O'Hanlon D, Kerin M, et al. Are elevated levels of the tumour marker CA19-9 of any clinical significance?--an evaluation. Ir J Med Sci 1999;168:124-6. [Crossref] [PubMed]
- Pokorny RM, Hunt L, Galandiuk S. What's new with tumor markers for colorectal cancer? Dig Surg 2000;17:209-15. [Crossref] [PubMed]
- Mackinnon RN, Selan C, Wall M, et al. The paradox of 20q11.21 amplification in a subset of cases of myeloid malignancy with chromosome 20 deletion. Genes Chromosomes Cancer 2010;49:998-1013. [Crossref] [PubMed]
- Sawayama H, Ishimoto T, Sugihara H, et al. Clinical impact of the Warburg effect in gastrointestinal cancer Int J Oncol 2014;45:1345-54. (review). [Crossref] [PubMed]
- Liu P, Chen H, Han L, et al. Expression and role of V1A subunit of V-ATPases in gastric cancer cells. Int J Clin Oncol 2015;20:725-35. [Crossref] [PubMed]
- Daniel C, Bell C, Burton C, et al. The role of proton dynamics in the development and maintenance of multidrug resistance in cancer. Biochim Biophys Acta 2013;1832:606-17. [Crossref] [PubMed]


