
Comprehensively analyze the expression and prognostic role for ten-eleven translocations (TETs) in acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease of the blood system characterized by the clonal expansion and differentiation arrest of myeloid progenitor cells. The incidence of AML increases by age, and the older patients, the more mortality (1). An estimated 19,940 new diagnostic AML patients, and 11,180 people die of AML in the United States (2). However, intensive chemotherapy is not a suitable option for many older patients with significant comorbidities, baseline organ dysfunction, or poor performance status, in whom the risk of complications and treatment-related mortality is unacceptably high. The worse thing is that relapsed AML patients only have less than 25% complete remission rate (3). Since the more efficient salvage regimens have been into practice, the complete response rate has increased, but these patients cannot obtain substantial CR duration (4.9 to 9.8 months) or survival (6.2 to 8.7 months) (4), this result did not be satisfied. Furthermore, the numbers of AML patients gradually increase in times. Thus, we urgently need more sensitive and specific markers for early diagnosis, more efficient but less side effect therapy for AML patients.
Although the advance of cell biology and comprehensive genomic analyses has shown the possible leukemogenesis mechanisms, it is still incompletely understood. The genes aberrations have been confirmed, one of the essential leukemogenesis drivers, even can as biomarkers indicated the favorable or adverse outcome of AML patients. Many studies showed that some improved cytogenetic changes play crucial roles in tumorigenesis and prognosis of AML, including aberration of TP53 (5), WT1 (6), double CEBPA mutation (7), RUNX1 (8), DNMT3A (9), FLT3 (10), and so on. DNA’s methylation has appeared relatively stable epigenetic process. However, with recent studies improve the function of ten-eleven translocation (TET) family proteins as 5-methylcytosine oxidases, the view has been reversed. The TET family, including TET1, TET2, and TET3. Over the last decade, TET protein or family functions and differential expression in various types of cancers have further insight. Many studies explore the TETs role in different cancers. For solid tumors, TET1 accelerates the triple-negative breast cancer exacerbation (11), but TET2 as inhibitors tumorigenesis of breast cancer cells in another study (12). In ovarian cancer, high expression of TET3 as an adverse biomarker (13). About hematological malignancies, TET1 mutations have been observed in chronic lymphocytic leukemia (CLL) (14) and T-cell acute lymphoblastic leukemia (T-ALL) (15), and AML (16). TET2 mutations also were found in MDS (17) and AML (18). However, TET3 changes less be found in hematological malignancies. In different cancer types, the TETs functions may be different. For hematological cancers, promoting DNA demethylation and regulate immunity is the most important biological function of TETs (19). Significant gene methylation, immunity tolerance, and immunity evasion significantly influence tumorigenesis, therapy resistance, and tumor progress. From these, we use public databases to analyse the prognostic value of TETs in AML and observing whether there are significant expression levels between AML samples and normal samples. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3149).
Methods
Expression analysis
The Oncomine database (20) and GEPIA (Gene Expression Profiling Interactive Analysis) (21) were preliminarily used to analyze the TETs expression levels between tumor samples and normal samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Analyzing the aberrations types, location of mutations, and structure of TETs
We used cBioPortal to learn the types of aberrations of TETs, perform the aberration locations, and construct the three-dimension structure of TETs (22).
Survival analysis
To investigate the relation TETs expression levels with the prognosis of AML patients, we use the UALCAN to analyze the TCGA data of AML patients (23). Further, we use GenomicScape (http://www.genomicscape.com) that data (GSE12417) derived from the GEO database to validate the survival results of AML (24). Also, we analyze the significantly correlated genes prognostic value of AML. Finally, we explore the relation TETs methylation with the prognosis of AML patients via METHsurv (25).
GeneMANIA analysis
GeneMANIA is a commonly used website for performing protein-protein interaction (PPI) network analysis and predicting the function of preferred genes (26). This user-friendly online tool can display gene or gene lists using bioinformatics methods, including gene co-expression, physical interaction, gene co-location, gene enrichment analysis, and website prediction. We predicted the TETs family’s function and significantly correlated genes and visualizing the gene networks via GeneMANIA.
Constructing the miRNA networks
We search for the miRNAs that target TETs via miTarbase (27), miRTarBase (27), and miWalk (28) in various databases to find out. Further, we performed the Venn diagram to find the TETs’ co-targets miRNA.
LinkOmics and ChEA3 analysis
The LinkedOmics database (http://www.linkedomics.org/login.php) is a Web-based platform for analyzing 32 TCGA cancer-associated multi-dimensional datasets (29). Data from the LinkFinder results were signed and ranked, and GSEA was used to analyze GO (CC, BP, and MF), KEGG pathways, and kinase-target enrichment. We used LinkOmics to analyze the CC, BP, and MF), kinase-target enrichment, and the significantly correlated genes of TETs. Further, we used the ChEA3 to find the TETs’ significant transcription factors (TFs) annotating the potential biological functions, and constructing the network of top 10 TFs (29).
Analyzing the cancer pathway activity and drugs sensitivity of defined genes
Cancer Analysis (GSCALite) analyzes Gene expression associated cancer pathway activity and Drug sensitivity for genes (30). We used the GSCALite dataset to explore the role of defined genes in the cancer pathway, further investigate the drug sensitivity of defined genes.
Validation of mutated types of TETs in AML patients
We selected the AML patients with TETs mutations in The Department of Hematology, the First Affiliated Hospital of Guangxi Medical University, to find the mutated types of TETs in our department.
Statistical analysis
The gene expression, survival analysis, and the protein-protein network relations P value <0.05 is considered a significant difference in our study.
Results
Transcriptional expression levels of TETs in AML patients
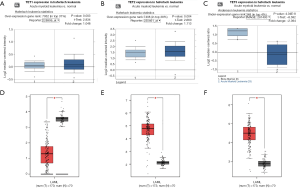
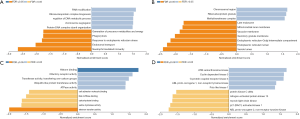
To identify the differential expression levels of TETs in AML and normal samples, the TETs mRNA levels in leukemia samples and control samples were analyzed by the Oncomine (Figure 1A,B,C) and GEPIA (Figure 1D,E,F) databases. The results of Oncomine analysis showed that the expression of TET1 (P=0.003) and TET2 (P=0.004) was significant overexpression in AML patients in Haferlach leukemia, but the TET3 (P=4.04e-8) was downregulation in Andersson leukemia samples. To further validate to TETs expression level, we used the GEPIA to perform analysis. The results indicated that TET2 and TET3 were significantly upregulated expression, but TET1 was significantly downregulated expression.
Aberrations types and the structure of TETs
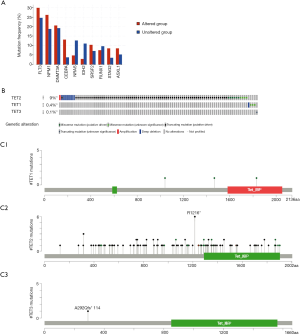
To investigate the aberrations types, aberrations locations, 3D structure of TETs, we used the C-BioPortal database to analyze. As the results have shown the top 10 co-expression of TETs including FLT3, NPM1, DNMT3A, CEBPA, NRAS, IDH2, SRSF2, RUNX1, STAG2, and ASXL1 (Figure 2A). The most standard mutated types of TET2 and TET1 were deep deletion and missense mutations (Figure 2B). The locations of TETs were shown in Figure 2C.
The relation between TETs expression levels and the prognosis of AML patients
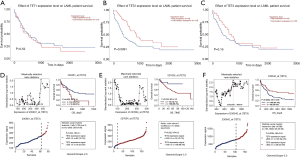
To understand whether the expression levels of TETs would have a significant influence on AML patients’ survival, we used the UACLAN database to clarify the relation between TETs expression and prognostic role of AML. As the results have shown, the higher expression TET2 may lead to positive outcomes of AML patients (P=0.0091). However, TET1 and TET3 expression levels did not impact the prognosis of AML patients (Figure 3A,B,C). To further validate the impact of TETs expressions for AML patients, we used the GSE12417 data to analyze. The results showed that high expression TET2 leads to poor survival for CN-AML patients, but high expression TET3 indicated a protective factor (Figure 3D,E,F).
Prognostics role of significantly correlated genes in AML
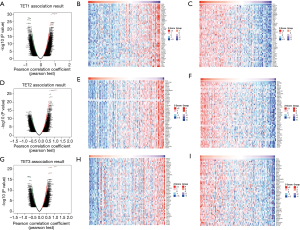
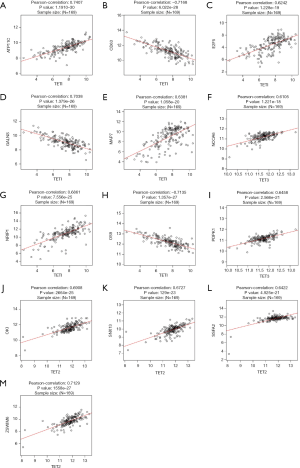
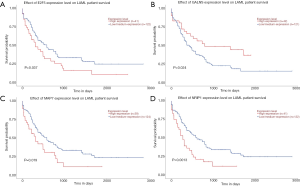
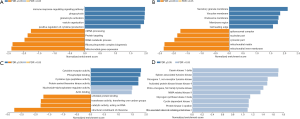
To further investigate the potential role of differentially expressed TETs in AML, the LinkFinder module of LinkedOmics was used to analyze mRNA sequencing data from 173 patients in TCGA. In the volcano plot, the dark red dots show a significantly positive correlation with TET1, TET2, and TET3, whereas dark green dots show a significantly negative correlation (false discovery rate, FDR <0.01). The top 50 significant gene sets positively and negatively associated with TET1, TET2, TET3, as shown in the heat maps (Figure 4A,B,C,D,E,F,G,H,I). Meanwhile, the statistical scatter plots show the association TETs expression with significantly correlated genes (Pearson’s correlation ≥0.6). From the results of association genes, we can observe APT11C, CD63, OS9, GALNS, NRIP1, MAP7, and E2F5 were strong associations with TET1; ZSWIM6, QKI, SNX13, and SSFA2 were strong associations with TET2; PDPK1 and NCOA6 were strong associations with TET3 (Figure 5A,B,C,D,E,F,G,H,I,J,K,L,M). To explore whether the strong association with TETs genes that have a significant impact on AML patients, we used UALCAN to analyze the TET1, TET2, TET3 strong related genes in OS of AML patients. There were four correlated significant genes have a significant impact on the OS of AML patients, the high expression GALNS (P=0.024) leads to an adverse outcome, the downregulation of E2F5 (P=0.037), MAP7 (P=0.019), and NRIP1(P=0.0013) indicated the excellent prognosis (Figure 6A,B,C,D).
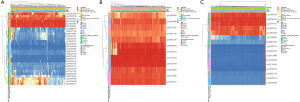
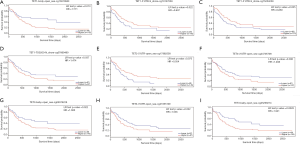
The relationship TETs methylation with the prognosis of AML
From the important role of methylation in tumorigenesis and prognosis of AML patients, we used METHsurv to analyze the TETs methylation status and the association with prognosis for AML patients, respectively. The locations of TETs methylation were shown in the heatmap (Figure 7A,B,C) and the prognosis of TETs methylation for AML patients was shown in Figure 8A,B,C,D,E,F,G,H,I. The hypermethylation of TET1 including cg19127638, cg12548760, cg18515801, cg01093854, cg13810683, and cg27426824, and the hypermethylation of TET2 including cg20586654, cg08530497, cg22794775, cg09666717, and cg17862558. TET3 has the most frequency of hypermethylation including cg02956499, cg13808088, cg00755592, cg15827185, cg21855109, cg05976228, cg25299214, cg17213010, cg24705708, cg02237855, cg01244346, and cg01355757 (Figure 7A,B,C). Further, we analyzed the prognostics value of TETs methylation in AML patients. The results indicated that hypermethylation of cg24705708 (P=0.036), cg05976228 (P=0.022), cg21855109(P=0.022), and cg25299214 (P=0.0028), cg17862558 (P=0.0073), cg13810683 (P=0.013), cg19127638 (P=0.022), cg15254238 (P=0.025), and cg07669489 (P=0.037) were the adverse biomarkers for AML patients (Figure 8A,B,C,D,E,F,G,H,I).
Functional enrichment analysis of TETs
To investigate the potential functional enrichment among TETs, we used the LinkedOmics database to analyze the significant enriched GO biological process (BP), cellular component (CC), molecular functions (MF), and target kinase. The main enrichments result in DNA transcription, RNA production, energy metabolism pathway, proteolysis, angiogenesis, and so on (Figures 9-11). The significant kinase targets of TET1 were CDK2 and TGFBR2. For TET2, there were CSNK1D, IRAK4, SYK, EGFR, MAP3K4, and NEK6 kinase targets. TET3 has the most numbers of kinase targets that include MYLK3, MYLK4, SGK1, CSNK1D, PLK2, CDK2, ILK, PRKCZ, PRKCE, MAPK10, MYLK2, and MYLK. Among the kinase targets, CDK2 was the TET1 and TET3 kinase co-targets, and CSNK1D was the kinase co-targets of TET2 and TET3 (Figures 9D,10D,11D).

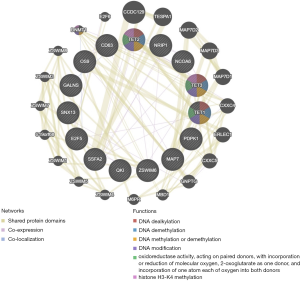
Further, the protein-protein network of TETs and the significantly correlated genes was constructed via GeneMANIA. Both of TETs interacted with CXXC4, but only TET3 and TET1 interacted with DNMT1, there still were many other significant interactions. The PPI network showed that the functions of TETs were covalent chromatin modification, demethylation, histone modification, histone H3-K4 methylation, histone lysine methylation, and DNA methylation or demethylation (Figure 12).
The regulated miRNA network of TETs
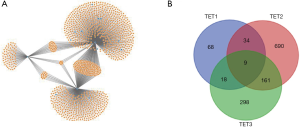
We used miTarbase, TargetScan, and miWalk databases to find the regulated miRNA of TETs, there were 690, 298, and 68 regulated miRNAs for TET2, TET1, and TET3, respectively. The network of TETs and miRNAs was constructed by miWalk (Figure 13A). There were 9 miRNAs including hsa-miR-2278, hsa-miR-105-3p, hsa-miR-6882-5p, hsa-miR-4732-3p, hsa-miR-4524a-5p, hsa-miR-6866-5p, hsa-miR-5580-3p, hsa-miR-4692, and hsa-miR-649 that their co-target genes were TETs (Figure 13B).
TF targets of TETs
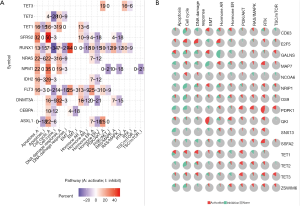
Further, we used the ChEA3 to analyze the TF targets of TET1, TET2, and TET3. Also, the analysis results of TFs indicated TFs of TETs were scattered in the nervous system, digestive system, and peripheral blood, further enrichment TFs functions they are the regulators of the immune response, cell differentiation, organ development, DNA transcription, and so on (Figure 14A,B). Further, we construct the encode TFs network and show the top 10 rank TFs, and the top 10 TFs of TETs are JUN, FOSL1, ZNF263, CTCF, MYOD1, TCF12, SP1, JUND, ETS1, and NFIC (Figure 14C,D). From these biological functions of kinase targets and TFs, the TETs may play essential roles in tumorigenesis and prognosis.
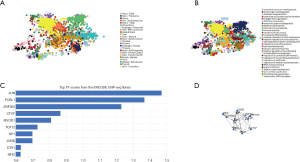
Cancer pathway activity and drugs sensitivity of TETs and correlated genes
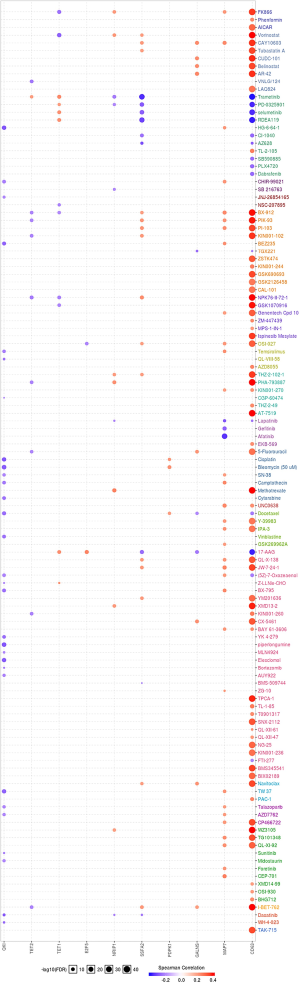
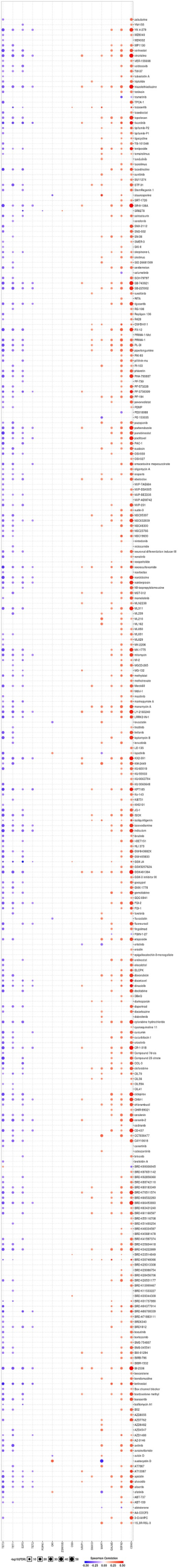
To incite the role of defined genes in the cancer pathway, the GSCAlite was used to perform the cancer pathway activity, and further investigate whether have drugs target the genes. The cancer pathway activity indicated TETs activated the PI3K/AKT signal pathway, but have a bidirectional function (activation and inhibition) in the DNA repair response, receptor tyrosine kinase (RTK), and cell cycle. The significantly correlated genes have an essential role in these cancer pathways as well, including GALNS, MAP7, and ZSWIM6 as the activators in TSC/mTOR; NCOA6, NRIP1, SNX13 are the inhibitors of apoptosis, and other genes also as the crucial parts in these cancer pathways (Figure 15A,B). The final goals of much cancer research are that it can provide useful information for therapying cancer patients, so we further analyze the potential drug targets of these defined genes. We used GSCAlite to construct the sensitivity of the drug of these genes that drugs sensitivity data from Cancer Therapeutics Response Portal (CTSP) and Genomics of Drug Sensitivity in Cancer (GDSC). The Spearman correlation represents the gene expression correlates with the drug. The positive correlation means the gene’s high expression is resistant to the drug. The results from GDSC shown trametinib was the most resistant for TET1 and TET2, but vorinostat and VNLG/124 were the most sensitive of TET1 and TET2, respectively. CD63 has the largest numbers of resistant drugs, including navitoclax, GSK1070916, belinostat, tubastatin A, vorinostat, phenformin, but QKI has the most numbers of sensitive drugs including TW37, piperlongumine, bortezomib, and bleomycin (Figure 16). Besides, the results of drug sensitivity from CTSP indicated they have many sensitive drugs for TETs, including alisertib, alvocidib, AT13387, belinostat, BI-2536, ceranib-2, CR-131-B, docetaxel, tivantinib, vincristine, but CD63 has the most significant number of resistant drugs (Figure 17).
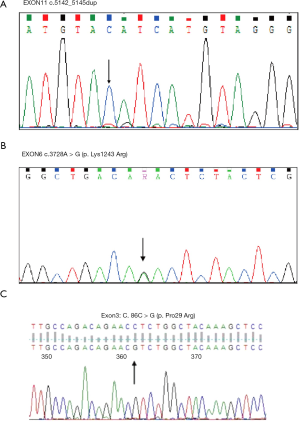
The mutated types of TETs in our department
We selected the AML patients in the Department of Hematology, the First Affiliated Hospital of Guangxi Medical University from January 2019 to January 2020. It only had three patients with TET2 mutations—also, the mutated type, including missense and truncating mutation (Figure 18A,B,C).
Discussion
Many cancers have aberrant promoter methylation, and it is considered to have a significant relation with tumorigenesis. For recent studies, TETs have seemed crucial regulators of the DNA demethylation pathway in various cancers. The functions of TET2 aberrant mutations in AML have been well elaborated; the TET2 gene has a relationship with leukemogenesis, therapy response, and prognosis. However, the other member of TETs’ (TET1 and TET3) role in AML remains unclear. Thus, we used bioinformatics to investigate the TETs expression levels, prognostics values, and the potential biological functions in AML.
The aberrant mutations of TETs not only have been studied in hematological cancer but also solid tumors, including breast cancers (11), prostate cancer (31), colon cancer (32), and melanoma (33). TET1 was an activator in TNBC by upregulating the oncogenic signaling, and the high TET1 expression level may lead the drug resistance by targeting the PI3K-mTOR pathway (11), so the high expression TET1 may indicate the adverse prognosis of TNBC. The research observed that TET1 has a relationship with cancer patients’ immunity regulating (34). To date, TET2 has seemed like the one that closest interaction with hematological malignancies. Cimmino et al. revealed TET2 as a tumor suppressor in leukemia, the restoration of the TET2 function could inhibit the cancer cell self-renewal and leukemia progression (35).
Further, Xu et al. study showed that activated the TET2 may improve the efficacy of immunotherapy efficacy and enhance the immunity of solid cancer patients (36). According to TET3, Cao et al. research showed that TET3 high expression indicated the poor prognosis in ovarian cancer patients (13). However, Ye et al. revealed TET3 is a tumor suppressor of ovarian cancer through inhibiting epithelial-mesenchymal transition. These studies revealed that the expression of TETs could be detectable in multiple cancer types, and the functions of TETs may be different in different cancer types or even subtype of the same cancer.
In this study, TETs expression levels showed TET1 significantly decreased expression, but the TET2 and TET3 were significant upregulation in AML samples via GEPIA analysis. The Oncomine analysis showed that TET1 and TET2 were significant overexpression in Haferlach Leukemia, but TET3 was a significant low expression in Andersson Leukemia. The TETs subgroups analyses indicated that TETs have significant differential expression levels between different subtypes, age, and other clinical features, these results may be one reason to explain the TETs different expression in various leukemia samples. Some studies approved the expression of TET1 and TET2 can impact the prognosis of AML patients. In our study, the OS results showed that only the TET2 expression level as a significant factor for AML prognosis via UALCAN, and the TET2 high expression leads to positive outcomes of AML patients.
Further, the validation OS results indicated TET2 and TET3 expression levels have a significant relation with AML prognosis, the high expression of TET2 indicating the poor prognosis, but the high expression levels of TET3 may be the protective factor for AML patients. One of the reasons for explaining this is that the validation of AML patients is the cytogenetically normal. These results indicated that the TET2 and TET3 expression levels might have different functions in different AML subtypes. Few studies investigated the relation between TET3 and AML. So, the underlying mechanisms of TETs in leukemogenesis need to be further explored.
Many studies approve DNA methylation as a critical part of tumorigenesis, progress, and metastasis in solid tumors. Emerging evidence reveals that the downregulation of DNA methylation may be a critical event for leukemogenesis and progression (37). TET one of the main roles is demethylation, but few studies explored the association TET methylation with the prognosis of AML. Herein, this study showed that some located TETs methylations significantly impact AML patients’ outcomes.
The regulation of TETs in AML further interested us. We constructed the gene interaction network of TETs and explored their GO enrichment and KEGG pathway analysis. As expected, these genes’ functions were primarily related to covalent chromatin modification, demethylation, histone modification, histone H3-K4 methylation, histone lysine methylation, and DNA methylation or demethylation.
These results indicated that the TETs as the vital regulator of DNA transcription and methylation. Both of these biological functions that take part in the tumorigenesis, progress, and metastasis. These results provide more information to explore further the TET’s biological role in leukemogenesis and progression.
Dysregulation of miRNAs has been found in multiple cancers; miRNAs can act as promoters or inhibitors of tumors. Hematologic malignancies are no exception; the miRNA’s aberrant expression may lead to hematologic cancer, different hematopoietic lineages. Distinctive miRNA profiles were observed in multiple cytogenetic subtypes of AML, and the miRNA expression levels were associated with the prognosis of AML (38). In our study, we finally acquired nine miRNAs that targeted the TETs. Kaymaz et al. revealed the has-miR-2278 act as the tumor suppressor; high expression miR-2278 can inhibit leukemic cell proliferation and induce apoptosis (39). To date, there are no studies that explore the other eight miRNAs’ role in any disease.
Tumorigenesis involves the changes of multi-omics that include gene aberration, changes of TF and the kinases, and so on. Single gene aberration may not be attributed to cancer. So, we find the significantly correlated genes of TETs, exploring their association and investigating the prognostic role in AML patients, finding the significant TFs and kinases, and incite their functions. For TETs, there are four significant correlated genes (GALNS, E2F5, MAP7, and NRIP1) that significantly impact the prognosis of AML patients. High expression GALNS indicated the poor outcomes of AML patients, downregulation of E2F5, MAP7, and NRIP1 as the protective factors of AML patients. Ho et al. study approved GALNS can as the diagnostics biomarker of multiple cancers (40), but no study of GALNS in AML. Aberrational E2F5 expression has been observed in various cancers. Kothandaraman et al. study revealed the E2F5 could act as a biomarker for diagnosing epithelial ovarian cancer (41). A study showed that E2F5 accelerated prostate cancer cell migration and invasion by regulating TFPI2, MMP-2, and MMP-9 (42). Many studies result revealed that targeted E2F5 can inhibit cancer development by various miRNAs (43-45). Research indicated that the E2F5 upregulation in AML, but the E2F5 function in AML, did not further be explored. The BP of E2F5 is related to cell cycle progression and transformation, and acting downstream of the TGF pathway (46). Aberrant, the changes of MAP7 have been observed in multiple cancers as well. Zhang et al. revealed MAP7 corporates with RC3H1 as the regulator of cell-cycle progression in cervical cancer cells by activating NF-κB signaling (47).
Further, Zhang et al. study showed MAP7 was a promotor of migration and invasion in cervical cancer by regulating autophagy (48), this result indicated the high expression MAP7 might lead to a poor prognosis for the patients with cervical cancer. According to AML, a study observed that high expression MAP7 indicated the short OS of young patients with CN-AML (49). There was a study that found that suppressed NRIP1 can inhibit breast cancer growth and cell-induced apoptosis (50). Both of the selected correlated significant genes indicate that high expression as an adverse biomarker for AML. However, the role of the selected significant genes in AML and the relation with TETs still unclear. It is needed to be further investigated.
The kinases play critical roles in gene expression and signaling transduction. For TETs, there are so many significant kinases, including CDK2, TGFBR2, CSNK1D, IRAK4, SYK, EGFR, MAP3K4, MYLK3, MYLK4, SGK1, PLK2, ILK, PRKCZ, PRKCE, MAPK10, MYLK2, and MYLK. Their main functions are the regulators of the cell cycle, cell growth, energy metabolism. Among the kinases, only CDK2 has a significant relation with AML. Shao et al. research showed that inhibiting the CDK2 can enhance all-trans-retinoic acid efficacy in AML cells (51). Further, Ran et al. also revealed CDK2 interacted with CR6-interacting factor 1 (CRIF1) to inhibit the leukemia cell cycle (52). The function of CDK2 is a regulator of the cell cycle. Dysregulation of the cell cycle is an essential part of tumorigenesis and drug resistance for some cancer types. Some research has already investigated the anticancer agents, targeting therapy the cell cycle (53,54). Regrettably, no study investigates the other kinases in AML. However, most of these kinases work as the promotor in various solid cancers, such as MYLK, promoting the migration and invasion of bladder cancer (55) and gastric cancer (56). PRKC as the accelerator of colon cancer (57). Inhibiting CSNK1D can weaken the migration and invasion ability of TNBC (58), and so on. These studies indicated that most of these kinases might act as an essential role in tumorigenesis and prognosis.
Many factors regulate gene expression; the TFs may be one of the most critical regulatory factors because most regulators impact gene expression by directly or indirectly affecting TFs. Our study used ChEA3 to find the TETs’ TFs and annotated them. The GO enrichment of ENCODE TFs revealed they are the regulators of DNA and RNA production. Among the TFs, Jun is the one most significantly related to TETs. Most of these TFs play a key role in different cancer types. Aberrant jun (including c-jun, jun-B, and jun-D) expression has been observed in multiple cancers. Activation of C-Jun can promote liver cancer development and sorafenib resistance (59). The expression of jun has been observed in AML, as well as. Inhibiting the jun can induce the death of leukemia cells by regulating unfolded protein response (60). The ETS1 proto-oncoprotein is a member of the Ets family of transcription factors that share a unique DNA binding domain, the ETS domain. ETS1 regulates the leukemic progenitor cell by effecting the autocrine GM-CSF transcription (61). Sp1 has a significant relation with drug resistance in AML by regulating the ERK-MSK MAPK signaling pathway to impact the survivin expression (62). For this, high expression Sp1 may lead to the poor outcome of AML. The other top TFs do not have any study related to AML, but most of them serve as the promotor of tumorigenesis and progression in multiple solid cancers. These clues indicate that the related TFs of TETs also play a key role in AML as well.
All cancer research aims to provide the underlying mechanisms of tumorigenesis, a biomarker of diagnosis and prognosis, and even find the new therapy target for cancer patients. Thus, patients with cancer can lengthen their lifespan, even cure the disease. Most cancer-related gene aberrant mutations act as the critical parts of multiple cancers’ tumorigenesis and tumor progression via the cancer pathway. In the present study, we used the GSCAlite to explore the role of TETs and the significantly correlated genes in cancer pathways, further investigating the potential drug targets of these genes. Among the cancer pathway, the PI3K/AKT signaling pathway has a significant relation with drug resistance in AML (63).
Furthermore, the PI3K/AKT signaling pathway is the regulator of leukemic cell fate that inhibits it can promote leukemic cell apoptosis (64). From these, a study explored the efficacy of targeting the PI3K/Akt signaling pathway in AML (65). Since the TETs are the positive regulator of the PI3K/Akt signaling pathway, we may conclude TETs are the promotor for AML. In AML, the oncofusion proteins, self-renewal activity, and leukemogenic program in hematopoietic cells have a significant association with a dysregulation in the DNA repair pathway, the accumulation of DNA damage that seems like the oncogenic driver of AML. Through these mechanisms, the DNA repair pathway may be the novel potential therapeutic target of AML. Yang et al. revealed that combinatorial therapy with AT-101 and IDA could eliminate leukemia stem-like cells via blockage of DNA damage repair (66). In our study, TET1 as activator but TET2 and TET3 have a bidirectional role in the DNA repair pathway.
Further research should focus on the underlying relation TET2 and TET3 with DNA repair response. About 40–60% AML patients with receptor tyrosine kinases (RTKs) or their downstream effector’s mutations (67,68). Among the RTKs, FLT3 and C-KIT have been well explored in AML. Most RTKs indicate the poor prognosis of AML. The inhibitors of FLT3 (69) and c-KIT (70) have been introduced to the practice. Both of these TETs acts as the activator or inhibitor in the RTK pathway. So, the TETs may have a different role in different AML types. The TSC/mTOR signaling pathway’s role is not unclear in cancer, the primary function of the TSC/mTOR signaling pathway is that it regulates the inflammatory responses (71), but there is not cancer research. Apoptosis is one of the essential types of cancer cell death. Many chemotherapy drugs achieve efficacy by inducing cancer cell apoptosis. The significantly correlated genes that NCOA6, NRIP1, and SNX13 inhibit the apoptosis. These genes may be as possibly a factor for the drug resistance of cancer patients. However, no study investigated the functions of NCOA6 and SNX13 in cancer. For the vital role of TETs and the significantly correlated genes in the cancer pathway, we use GSCAlite to explore them’ potential target drugs. Finally, we found the alisertib, alvocidib, AT13387, belinostat, BI-2536, ceranib-2, CR-131-B, docetaxel, tivantinib, and vincristine are the drugs that TETs sensitive. These results can provide useful information for the clinician to choose the drugs for therapy, TETs mutation, or correlate significant AML patients’ genes.
Conclusions
In the present study, we comprehensively analyze the TET’s expression levels, prognostics role, methylation, biological functions with AML patients. Also, further explored the significant TFs, kinases, cancer pathways, and drug targets of TETs. Our results suggest that TET1, TET2, and TET3 were differentially expressed in AML, CN-AML patients were benefited from overexpression of TET3, but overexpression of TET2 leads to adverse prognosis for CN-AML and other AML patients. The hypermethylation cg24705708, cg05976228, cg21855109, cg25299214, cg17862558, cg13810683, cg1912763, cg15254238, and cg07669489 of TETs were the adverse biomarkers for AML patients. We found that significantly correlated genes, including GALNS, E2F5, MAP7, and NRIP1, can be the novel prognostic biomarker for AML. Regulatory network analysis suggested TETs regulate covalent chromatin modification, demethylation, histone modification, histone H3-K4 methylation, cell cycle, and immune response through pathways involving the significant kinases, TFs, and miRNA. Finally, cancer pathway analysis of TETs activates the PI3K/AKT signal pathway, but have a bidirectional function (activation and inhibition) in the DNA repair response, receptor tyrosine kinase (RTK), and cell cycle. Drug sensitivity analysis results reveal that trametinib was the most resistant for TET1 and TET2, vorinostat, and VNLG/124 were the most sensitive of TET1 and TET2 by GDSC, respectively. Both TETs are sensitive for alisertib, alvocidib, AT13387, belinostat, BI-2536, ceranib-2, CR-131-B, docetaxel, tivantinib, vincristine by CTSP.
Acknowledgments
We would like to thank the Oncomine and LinkedOmics databases for open access to the AML sequencing datasets.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3149
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3149). The authors have no conflicts of interest to declare.
(English language editor: J. Chapnick)
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400-4. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. 2020;70:7-30.
- Hu N, Cheng Z, Pang Y, et al. Prognostic effect of allogeneic hematopoietic stem cell transplantation on first and non-first complete remission in acute myeloid leukemia. Ann Transl Med 2019;7:500. [Crossref] [PubMed]
- Megías-Vericat JE, Martínez-Cuadrón D, Sanz M, et al. Salvage regimens using conventional chemotherapy agents for relapsed/refractory adult AML patients: a systematic literature review. Ann Hematol 2018;97:1115-53. [Crossref] [PubMed]
- Barbosa K, Li S, Adams PD, et al. The role of TP53 in acute myeloid leukemia: Challenges and opportunities. Genes Chromosomes Cancer 2019;58:875-88. [Crossref] [PubMed]
- Wang Y, Xiao M, Chen X, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 2015;57:662-73. [Crossref] [PubMed]
- Mannelli F, Ponziani V, Bencini S, et al. CEBPA-double-mutated acute myeloid leukemia displays a unique phenotypic profile: a reliable screening method and insight into biological features. Haematologica 2017;102:529-40. [Crossref] [PubMed]
- Jalili M, Yaghmaie M, Ahmadvand M, et al. Prognostic Value of RUNX1 Mutations in AML: A Meta-Analysis. Asian Pac J Cancer Prev 2018;19:325-9. [PubMed]
- Brunetti L, Gundry MC, Goodell MA. DNMT3A in Leukemia. Cold Spring Harb Perspect Med 2017;7:a030320. [Crossref] [PubMed]
- Smith CC. The growing landscape of FLT3 inhibition in AML. Hematology Am Soc Hematol Educ Program 2019;2019:539-47. [Crossref] [PubMed]
- Good CR, Panjarian S, Kelly AD, et al. TET1-Mediated Hypomethylation Activates Oncogenic Signaling in Triple-Negative Breast Cancer. Cancer Res 2018;78:4126-37. [Crossref] [PubMed]
- Zhu X, Li S. TET2 inhibits tumorigenesis of breast cancer cells by regulating caspase-4. Sci Rep 2018;8:16167. [Crossref] [PubMed]
- Cao T, Pan W, Sun X, et al. Increased expression of TET3 predicts unfavorable prognosis in patients with ovarian cancer-a bioinformatics integrative analysis. J Ovarian Res 2019;12:101. [Crossref] [PubMed]
- Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2011;44:47-52. [Crossref] [PubMed]
- Kalender Atak Z, De Keersmaecker K, Gianfelici V, et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS One 2012;7:e38463. [Crossref] [PubMed]
- Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood 2012;120:e83-92. [Crossref] [PubMed]
- Hirsch CM, Nazha A, Kneen K, et al. Consequences of mutant TET2 on clonality and subclonal hierarchy. Leukemia 2018;32:1751-61. [Crossref] [PubMed]
- Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009;360:2289-301. [Crossref] [PubMed]
- Feng Y, Li X, Cassady K, et al. TET2 Function in Hematopoietic Malignancies, Immune Regulation, and DNA Repair. Front Oncol 2019;9:210. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Kassambara A, Rème T, Jourdan M, et al. GenomicScape: an easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput Biol 2015;11:e1004077. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 2011;39:D163-9. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. [Crossref] [PubMed]
- Smeets E, Lynch AG, Prekovic S, et al. The role of TET-mediated DNA hydroxymethylation in prostate cancer. Mol Cell Endocrinol 2018;462:41-55. [Crossref] [PubMed]
- Huang Y, Wang G, Liang Z, et al. Loss of nuclear localization of TET2 in colorectal cancer. Clin Epigenetics 2016;8:9. [Crossref] [PubMed]
- Fu S, Wu H, Zhang H, et al. DNA methylation/hydroxymethylation in melanoma. Oncotarget 2017;8:78163-73. [Crossref] [PubMed]
- Collignon E, Canale A, Al Wardi C, et al. Immunity drives TET1 regulation in cancer through NF-κB. Sci Adv 2018;4:eaap7309. [Crossref] [PubMed]
- Cimmino L, Dolgalev I, Wang Y, et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 2017;170:1079-95.e20. [Crossref] [PubMed]
- Xu YP, Lv L, Liu Y, et al. Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J Clin Invest 2019;129:4316-31. [Crossref] [PubMed]
- Hsu PC, Kadlubar S, Su LJ, et al. County poverty levels influence genome-wide DNA methylation profiles in African American and European American women. Transl Cancer Res 2019;8:683-92. [Crossref]
- Ke S, Zhou X. LncRNA MVIH knockdown inhibits the malignancy progression through downregulating miR-505 mediated HMGB1 and CCNE2 in acute myeloid leukemia. Transl Cancer Res 2019;8:2526-34. [Crossref]
- Kaymaz BT, Günel NS, Ceyhan M, et al. Revealing genome-wide mRNA and microRNA expression patterns in leukemic cells highlighted "hsa-miR-2278" as a tumor suppressor for regain of chemotherapeutic imatinib response due to targeting STAT5A. Tumour Biol 2015;36:7915-27. [Crossref] [PubMed]
- Ho ML, Kuo WK, Chu LJ, et al. N-acetylgalactosamine-6-sulfatase (GALNS), Similar to Glycodelin, Is a Potential General Biomarker for Multiple Malignancies. Anticancer Res 2019;39:6317-24. [Crossref] [PubMed]
- Kothandaraman N, Bajic VB, Brendan PN, et al. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer 2010;10:64. [Crossref] [PubMed]
- Karmakar D, Maity J, Mondal P, et al. E2F5 promotes prostate cancer cell migration and invasion through regulation of TFPI2, MMP-2 and MMP-9. Carcinogenesis 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Tian H, Hou L, Xiong YM, et al. miR-132 targeting E2F5 suppresses cell proliferation, invasion, migration in ovarian cancer cells. Am J Transl Res 2016;8:1492-501. [PubMed]
- Li Q, Shi J, Xu X. MicroRNA-1271-5p inhibits the tumorigenesis of ovarian cancer through targeting E2F5 and negatively regulates the mTOR signaling pathway. Panminerva Med 2020; [Epub ahead of print]. [PubMed]
- Lin C, Hu Z, Yuan G, et al. MicroRNA-1179 inhibits the proliferation, migration and invasion of human pancreatic cancer cells by targeting E2F5. Chem Biol Interact 2018;291:65-71. [Crossref] [PubMed]
- Chen CR, Kang Y, Siegel PM, et al. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 2002;110:19-32. [Crossref] [PubMed]
- Zhang R, Li L, Chen L, et al. MAP7 interacts with RC3H1 and cooperatively regulate cell-cycle progression of cervical cancer cells via activating the NF-κB signaling. Biochem Biophys Res Commun 2020;527:56-63. [Crossref] [PubMed]
- Zhang L, Liu X, Song L, et al. MAP7 promotes migration and invasion and progression of human cervical cancer through modulating the autophagy. Cancer Cell Int 2020;20:17. [Crossref] [PubMed]
- Fu L, Fu H, Zhou L, et al. High expression of MAP7 predicts adverse prognosis in young patients with cytogenetically normal acute myeloid leukemia. Sci Rep 2016;6:34546. [Crossref] [PubMed]
- Aziz MH, Chen X, Zhang Q, et al. Suppressing NRIP1 inhibits growth of breast cancer cells in vitro and in vivo. Oncotarget 2015;6:39714-24. [Crossref] [PubMed]
- Shao X, Xiang S, Fu H, et al. CDK2 suppression synergizes with all-trans-retinoic acid to overcome the myeloid differentiation blockade of AML cells. Pharmacol Res 2020;151:104545. [Crossref] [PubMed]
- Ran Q, Hao P, Xiao Y, et al. CRIF1 interacting with CDK2 regulates bone marrow microenvironment-induced G0/G1 arrest of leukemia cells. PLoS One 2014;9:e85328. [Crossref] [PubMed]
- Tadesse S, Caldon EC, Tilley W, et al. Cyclin-Dependent Kinase 2 Inhibitors in Cancer Therapy: An Update. J Med Chem 2019;62:4233-51. [Crossref] [PubMed]
- Kollmann K, Briand C, Bellutti F, et al. The interplay of CDK4 and CDK6 in melanoma. Oncotarget 2019;10:1346-59. [Crossref] [PubMed]
- Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017;403:305-17. [Crossref] [PubMed]
- Xia N, Cui J, Zhu M, et al. Androgen receptor variant 12 promotes migration and invasion by regulating MYLK in gastric cancer. J Pathol 2019;248:304-15. [Crossref] [PubMed]
- Qureshi-Baig K, Kuhn D, Viry E, et al. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy 2020;16:1436-52. [Crossref] [PubMed]
- Bar I, Merhi A, Larbanoix L, et al. Silencing of casein kinase 1 delta reduces migration and metastasis of triple negative breast cancer cells. Oncotarget 2018;9:30821-36. [Crossref] [PubMed]
- Xiang DM, Sun W, Zhou T, et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut 2019;68:1858-71. [Crossref] [PubMed]
- Zhou C, Martinez E, Di Marcantonio D, et al. JUN is a key transcriptional regulator of the unfolded protein response in acute myeloid leukemia. Leukemia 2017;31:1196-205. [Crossref] [PubMed]
- Bade-Döding C, Göttmann W, Baigger A, et al. Autocrine GM-CSF transcription in the leukemic progenitor cell line KG1a is mediated by the transcription factor ETS1 and is negatively regulated through SECTM1 mediated ligation of CD7. Biochim Biophys Acta 2014;1840:1004-13. [Crossref] [PubMed]
- Zhang Y, Chen HX, Zhou SY, et al. Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Mol Cancer 2015;14:56. [Crossref] [PubMed]
- Liang H, Zheng QL, Fang P, et al. Targeting the PI3K/AKT pathway via GLI1 inhibition enhanced the drug sensitivity of acute myeloid leukemia cells. Sci Rep 2017;7:40361. [Crossref] [PubMed]
- Hao Y, Zhang N, Wei N, et al. Matrine induces apoptosis in acute myeloid leukemia cells by inhibiting the PI3K/Akt/mTOR signaling pathway. Oncol Lett 2019;18:2891-6. [Crossref] [PubMed]
- Bertacchini J, Heidari N, Mediani L, et al. Targeting PI3K/AKT/mTOR network for treatment of leukemia. Cell Mol Life Sci 2015;72:2337-47. [Crossref] [PubMed]
- Yang Q, Chen K, Zhang L, et al. Synthetic lethality of combined AT-101 with idarubicin in acute myeloid leukemia via blockade of DNA repair and activation of intrinsic apoptotic pathway. Cancer Lett 2019;461:31-43. [Crossref] [PubMed]
- Goemans BF, Zwaan CM, Miller M, et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia 2005;19:1536-42. [Crossref] [PubMed]
- Gari M, Goodeve A, Wilson G, et al. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol 1999;105:894-900. [Crossref] [PubMed]
- Zhao M, Kiyoi H, Yamamoto Y, et al. In vivo treatment of mutant FLT3-transformed murine leukemia with a tyrosine kinase inhibitor. Leukemia 2000;14:374-8. [Crossref] [PubMed]
- Longley BJ Jr, Metcalfe DD, Tharp M, et al. Activating and dominant inactivating c-KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci U S A 1999;96:1609-14. [Crossref] [PubMed]
- Weichhart T, Costantino G, Poglitsch M, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 2008;29:565-77. [Crossref] [PubMed]


