
Knockout of STK10 promotes the migration and invasion of cervical cancer cells
IntroductionOther Section
According to data from the World Health Organization, cervical cancer is the fourth most common cancer in women, with about 570,000 new cases in 2018, representing 6.6% of female cancers (https://www.who.int/cancer/prevention/diagnosis-screening/cervical-cancer/en/). It is the second leading cause of cancer-associated deaths in women, severely endangers women’s health, and imposes a substantial economic burden worldwide (1). Although significant advances in the diagnostic and therapeutic strategies of cervical cancer have been achieved in the past several decades, the overall 5-year survival rate for patients remains poor (2,3). The principal causes of the high mortality are metastasis and recurrence of cervical cancer (4). Therefore, it is necessary to clarify the underlying pathogenic mechanism and identify novel factors involved in the progression of cervical cancer, which is the only way to find new, effective methods for the prevention, diagnosis, and treatment of this disease.
STK10 (serine-threonine kinase 10), also called a lymphocyte-oriented kinase (LOK), is a serine/threonine kinase predominantly expressed in lymphoid organs. It has been demonstrated that STK10 can phosphorylate the conserved threonine residues at the C-terminal domain of ERM proteins (T567, T564, and T558, for ezrin, radixin, and moesin, respectively), which is essential for the activation of these proteins (5,6). ERM proteins are linking proteins that mediate the interaction between the actin cytoskeleton and the plasma membrane and are involved in several physiological processes such as maintenance of the cytoskeletal structure, cell motility, cell adhesion, cell movement, and signal transduction (7-9). Activated ERM proteins also play essential roles in the pathogenesis of various cancers via regulation of cytoskeletal rearrangement, cell migration, and invasion (10-12). STK10 is expressed in about 17 cancer types, including cervical cancer, according to the “Human Protein Atlas” (https://www.proteinatlas.org/ENSG00000072786-STK10). However, the biological functions of STK10 in the pathogenesis of cervical cancer remain undetermined.
In the present study, to investigate the function of STK10 in cervical cancer, we generated STK10 knockout (KO) HeLa and Caski cell lines using the CRISPR-Cas9 gene-editing system. We analyzed the effects of STK10 KO on tumor biological behaviors such as cell proliferation, apoptosis, adhesion, migration, and invasion.
MethodsOther Section
Cell culture
Cervical cancer cell lines (HeLa, Caski) were purchased from the American Type Culture Collection (USA) and cultured in DMEM and RPMI, respectively, with 10% (vol/vol) fetal bovine serum (FBS, Gibco) at 37 °C in a 5% CO2 incubator.
Plasmid construction
The guide-RNA (gRNA, 5-GGCGGACGTGCTCATATTCG-3¢) was designed by the online CRISPR design tool (https://zlab.bio/guide-design-resources). Two partially complementary oligonucleotides (5'-CACCGGCGGACGTGCTCATATTCG-3' and 5'-AAACCGAATATGAGCACGTCCGCC-3') were annealed and cloned into the PX459 plasmid (addgene, #62988) which was digested by the BbsI restriction enzyme. This construct was called the STK10 KO plasmid, which could guide hSpCas9 to the genomic target site in STK10.
Transfection and puromycin selection
A total of 4 µg STK10 KO plasmids and PX459 plasmids (control) were transfected into 8×105 cells with Lipofectamine 3000 reagent (Thermo Fisher), and the supernatant was exchanged for fresh medium 12 h later. After 2 days, the cells (including the control) were incubated in fresh medium with 1 µg/mL puromycin. After 7 days, cells resistant to puromycin could be selected. Western blot analysis was used to identify the effect of knockout. The HeLa STK10 KO and Caski STK10 KO cell lines that were not the single clone were established, and DNA sequencing by a specific sequencing primer (5'-GTGCTCCGAAACAGGGC-3') identified the genomic changes.
Western blot analysis
HeLa and Caski cell lysates were prepared using RIPA buffer [1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS in phosphate-buffered saline (PBS)] with freshly supplemented protease inhibitors cocktail (Roche). Next, the protein concentration was assayed by BCA (bicinchoninic acid) method, and 30 µg protein was separated on 10% SDS-PAGE membranes. Standard protocols were used for electrophoresis and immunoblotting analyses. Membranes were blocked with 5% milk in PBS. An anti-STK10 antibody (abcam, ab70484), anti-GAPDH rabbit polyclonal antibody (BBI, D110016), recombinant anti-ezrin antibody (abcam, ab40839), recombinant anti-moesin antibody (abcam, ab52490), recombinant anti-radixin antibody (abcam, ab52495) and Anti-ezrin (pThr567)/radixin (pThr564)/moesin (pThr558) antibody (abcam, ab76247) were used as the primary antibodies. The membranes were then incubated with IRDyeCW800-conjugated anti-rabbit immunoglobulin (LI-COR) and scanned with the LI-COR Odyssey imaging system (LI-COR).
CCK-8 cell viability assay
Cell viability was detected by cell counting Kit-8 (CCK-8; Dojindo Laboratories) assay. HeLa and Caski cells were seeded to a 96-well plate with 1,000 cells per well. Each group contained four duplicates. Ten percent CCK-8 assessed cell viability in a complete medium per 24 h. The absorbance was detected at 450 nm using a microplate reader (BioTek). The relative cell viability is the ratio of absorbance at the time relative to 0 h.
BrdU incorporation and apoptosis detection
Cell proliferation was detected by Phase-Flow FITC BrdU Kit (BioLegend). Firstly, the BrdU solution at 0.5 µL/mL was added to the cell suspension and cultured for 1 h. The cells were then dyed by anti-BrdU antibody and 7-AAD and analyzed by flow cytometry. The Annexin V-FITC Apoptosis Detection Kit (eBioscience) was used to detect apoptosis of cells according to the manufacturer’s instructions. HeLa and Caski cells were seeded to a 6-well plate with 5×105 cells per well. Each group contained three duplicates. The cells were digested with 0.25% trypsin after 24 h, dyed with fluorochrome-conjugated AnnexinV and propidium iodide (PI) staining solution and analyzed by flow cytometry.
Cell adhesion assay
The human plasma fibronectin (FN, Sigma-Aldrich) stock solution was diluted to 40 µg/mL in ddH2O. A 96-well culture plate was coated with 30 µL/well and incubated at 4 °C overnight. The plate was washed three times with PBS and blocked with PBS containing 1% bovine serum albumin at 37 °C in cell incubator for 1 h. After washing three times with PBS, 2×104 cells in 100 µL medium with 1% FBS were seeded to the well. Cells were incubated at 37 °C in a cell incubator for the appropriate time (HeLa: 30 min, Caski: 1 h). The wells were washed twice with PBS to remove unbound cells, and 100 µL medium with 1% FBS containing 10% CCK8 was added to each well, and the plate was incubated at 37 °C for 2 h. Absorbance was detected at 450 nm using a microplate reader (BioTek). Bound cells were fixed with 4% PFA, stained with 0.5% crystal violet for 5 min at room temperature, and then photographed at ×100.
Transwell assay
Cells were starved with serum-free medium for 24 h, and Transwell chambers with a pore size of 8.0 µm (Millipore) were inserted into a 24-well culture plate. For the cell invasion assay, the Transwell chambers were precoated with 80 µL Matrigel (Corning) for 2 h in a cell incubator, but not for the cell migration assay. Next, 5×104 HeLa cells or 1×105 Caski cells were respectively resuspended in 400 µL serum-free medium and added to the upper chamber, followed by the addition of 600 µL complete medium outside of the inserts and incubation at 37 °C in cell incubator for the appropriate time (HeLa: 24 h, Caski: 48 h). Cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet for 5 min at room temperature. All cells above the membrane were removed by cotton swab, and only the cells under the membrane were photographed at ×100. The number of cells in five random fields of view was counted to obtain an average sum of cells.
Wound healing assay
HeLa and Caski cells were seeded to a 6-well plate with 5×105 cells per well. When the cells reached 100% confluence, they were starved with medium containing 1% FBS for 24 h. A 200-µL pipette tip was used to create a wound through the center of the confluent cell layer, which was then washed twice with medium. Cells were treated with medium containing 1% FBS. Photographs of the wound were taken at 0, 24, 36, and 48 h. Image-J software was used to quantify the wound area. Wound healing percentage (area at 0 h – the area at the relevant time)/area of 0 h ×100%) was used to represent the cells’ migratory capacity.
RNA extraction and qRT-PCR analyses
RNA was extracted from the HeLa and Caski cell lines by RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s instructions, and 1 µg RNA was reverse transcribed to cDNA by AMV Reverse Transcriptase (Takara). The 1 µL cDNA product was used for quantitative real-time polymerase chain reaction (qRT-PCR) analyses with FastStart Universal SYBR Green Master (Roche) on an Eppendorf Mastercycler system according to the manufacturer’s instructions. All samples were tested in triplicate, and the results were normalized to GAPDH expression. Primers are listed below:
PCDHG-qRT-PCR-F: 5'-GAGATGCTGCAAGCCATGAT-3' PCDHG-qRT-PCR-R: 5'-CATTGCTGCCTGGGATGTAGA-3' MUC1-qRT-PCR-F: 5'-TATGAGCGAGTACCCCACCT-3' MUC1-qRT-PCR-R: 5¢-GCCACTGCTGGGTTTGTGTA-3' FOXF2-qRT-PCR-F: 5'-CCAGCATGTCCTCCTACTCG-3' FOXF2-qRT-PCR-R: 5'-TGGTGATAATACGACCCGCT-3' CDKN2A-qRT-PCR-F: 5'-ATCGCGATGTCGCACGGTA-3' CDKN2A-qRT-PCR-R: 5'-AATCGGGGATGTCTGAGGGA-3' CTNNB1-qRT-PCR-F: 5¢-GTGGACCACAAGCAGAGTGC-3¢ CTNNB1-qRT-PCR-R: 5'-CCTAAAGCTTGCATTCCACCA-3' GAPDH-qRT-PCR-F: 5'-TGACTTCAACAGCGACACCCA-3' GAPDH-qRT-PCR-R: 5'-CACCCTGTTGCTGTAGCCAAA-3'
RNA-Seq analysis
RNA-seq transcriptome libraries were prepared following the TruSeqTM RNA sample preparation Kit from Illumina, using 1 µg of total RNA. Messenger RNA was isolated with polyA selection by oligo (dT) beads and fragmented using fragmentation buffer. cDNA synthesis, end repair, A-base addition, and ligation of the Illumina-indexed adaptors were performed according to the manufacturer’s protocol. Libraries were then size-selected for cDNA target fragments of 200–300 base pairs (bp) on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After being quantified by TBS380, Paired-end libraries were sequenced with the Illumina HiSeq PE 2X151bp read length. The raw paired end reads were trimmed and quality controlled by Trimmomatic with default parameters (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic). Next, clean reads were separately aligned to the reference genome with orientation mode using Tophat (http://tophat.cbcb.umd.edu/) software (13). Tophat is a program that can align RNA-Seq reads to a genome in order to identify gene expression and exon-exon splice junctions. It is built on the ultrafast short read mapping program Bowtie2. We used the Tophat software to map with default parameters.
The expression level for each transcript was calculated using the fragments per kilobase of exon per million mapped reads (FRKM) method to identify DEGs (differential expression genes) between two different samples. Cuffdiff (http://cufflinks.cbcb.umd.edu/) was used for differential expression analysis (14). The DEGs between two samples were selected using the following criterion: the logarithmic fold change (FC) >2 and the FDR <0.05. Gene ontology (GO) functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) to understand the functions of the DEGs (15). DEGs were significantly enriched in GO terms and metabolic pathways when their Bonferroni-corrected P value was less than 0.05.
Statistical analysis
Each experiment was performed at least three times independently, and results are represented as the mean ± standard deviation (SD). All statistical analyses were performed using GraphPad Prism software (San Diego, CA, USA). Comparisons between two groups were performed using Student’s t-test. Differences were defined as statistically significant if P<0.05.
ResultsOther Section
Generation of STK10 KO cell lines with CRISPR-Cas9 gene-editing system
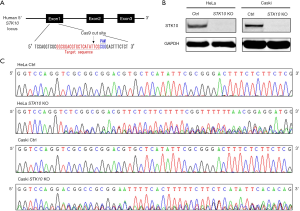
We used the CRISPR-Cas9 gene-editing system to generate STK10 KO cervical cancer cell lines. The oligonucleotides for the guide-RNA were designed, synthesized, and cloned into a pX459 vector (Figure 1A) for transfection transfected into HeLa and Caski cells. The indel mutations in these cell lines were confirmed by DNA sequencing of the PCR products of target DNA (Figure 1B). Finally, Western blot with anti-STK10 antibody showed that the expression of STK10 was abolished in the STK10 KO cell lines (Figure 1C).
Effects of STK10 KO on cervical cancer cell proliferation
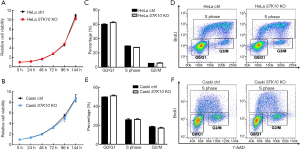
The effect of STK10 deficiency on the spontaneous growth of HeLa and Caski cells was first evaluated by CCK-8 assay from 1 to 6 days. As shown in Figure 2A,B, in either the HeLa or Caski cell line, the relative cell viability was comparable between STK10 KO and control cells. Furthermore, we conducted a BrdU incorporation assay and assessed the cell cycle distribution using fluorescence-activated cell sorter (FACS) analysis. The results indicated that the target deletion of STK10 did not affect the rate of BrdU incorporation in the cells. Moreover, no phases of the cell cycle were changed in the STK10 KO cell lines (Figure 2C,D,E,F). Our data revealed that STK10 is not necessary for cervical cancer cell proliferation or cell cycle progression.
Effects of STK10 KO on apoptosis of cervical cancer cells
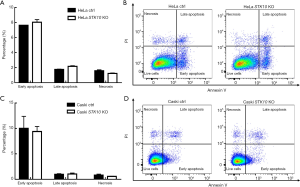
To study the effects of STK10 KO on apoptosis of cervical cancer cells, we performed AnnexinV/PI staining assay and flow cytometry analyses. As shown in Figure 3A,B,C,D, in both the HeLa and Caski cell lines, the proportions of early apoptotic cells (AnnexinV+PI−), late apoptotic cells (AnnexinV+PI+), and necrotic cells (AnnexinV−PI+) were comparable between STK10 KO and control cells. These data indicated that STK10 deficiency did not affect apoptosis of cervical cancer cells.
Depletion of STK10 and migration of cervical cancer cells
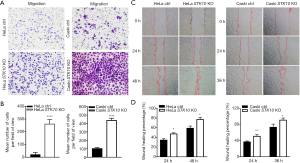
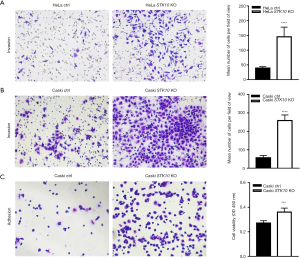
To explore the role of STK10 in the tumorigenesis and metastasis of cervical cancer, we assessed cell migration in vitro. The Transwell assay showed that the number of cells that migrated out increased in the STK10 KO group compared with the control group (P<0.0001; Figure 4A,B). As shown in Figure 4C,D, wound closure of the STK10-depleted cells accelerated. Collectively, these results suggested that the depletion of STK10 could promote the migration of cervical cancer cells and indicated that STK10 might play an essential role in the metastasis of cervical cancer.
Depletion of STK10 and invasion of cervical cancer cells
In order to better simulate the process of tumor invasion in vitro, we performed a Transwell assay with added Matrigel to detect whether STK10 affected cell invasion. The results showed that the STK10 KO cells had increased ability to migrate and invade across the Matrigel (by up to 80%) compared with control cells (P<0.0001; Figure 5A,B). Furthermore, the cell-matrix interaction was dramatically increased in the STK10 KO group (Figure 5C). These results suggested that the depletion of STK10 could promote the invasion and adhesion of cervical cancer cells, and further indicated that STK10 plays a vital role in tumor metastasis.
Regulation of the migration and invasion of cervical cancer cells by STK10
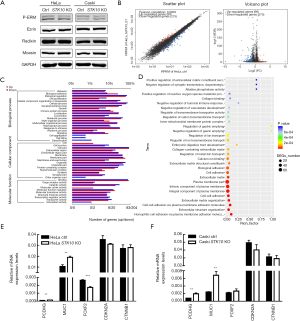
It is well established that STK10 is one of the kinases responsible for the phosphorylation of ezrin and other ERM proteins, which have been demonstrated to be essential in cell migration. The expression and phosphorylation levels of all three ERM-family proteins were comparable between STK10 KO and control cells (Figure 6A), which indicated that the effects of STK10 on cervical cancer cell migration and invasion are independent of ERM proteins.
Gene expression of cervical cancer cells lacking STK10
To investigate the mechanisms underlying the aberrant migration and invasion of STK10-deficient cells, we performed RNA-seq analysis for STK10 KO and the control group HeLa cells. Through comparison of the gene expression levels of the two genotypes, we identified 275 transcripts that were significantly decreased and 99 transcripts that were increased in STK10 KO HeLa cells based on FDR =0.05 & FC ≥2, as assessed by FRKM method using Cuffdiff software (Figure 6B). GO analysis of the differentially expressed transcripts further revealed that STK10 deficiency led to aberrant expression of the genes involved in various biological processes, including cell adhesion, extracellular matrix organization, and metabolic processes (Figure 6C,D). The expression levels of MUC1, PCDHG and some other DEGs that have been demonstrated to play critical roles in the migration and invasion of cancer cells were further confirmed using qRT-PCR in HeLa and Caski cells (Figure 6E,F). These findings implied that STK10 might execute various physiological functions through multiple pathways.
DiscussionOther Section
In this study, we assessed the role of STK10 in the development of cervical cancer using CRISPR-Cas9-mediated STK10 KO HeLa and Caski cervical cancer cell lines. The proliferation, apoptosis, migration, and invasive activity of these cells were respectively detected by BrdU incorporation, AnnexinV/PI staining, wound healing assay, and Transwell assays without and with Matrigel. The results showed that STK10 depletion does not affect cell proliferation or apoptosis, but promotes the adhesion, migration, and invasion of cervical cancer cells.
In recent years, genes related to tumor invasion have been receiving considerable attention among scientists. Cervical cancer is one of the most common malignant cancers that seriously threatens women’s health globally. It has been widely recognized that persistent infection with human papillomavirus (HPV) is one of the highest risk factors for cervical cancer (16-18). Early detection and prevention of HPV are effective against this malignant disease, but the effect of vaccination against HPV on the cervical cancer incidence will take several decades to emerge. Therefore, it is necessary to better understand the biology of this cancer in order to improve early diagnosis and treatment. STK10 is expressed in about 17 cancer types, including cervical cancer. Previous studies have shown that STK10 may affect LFA-1-mediated lymphocyte adhesion (19), while other studies report a mutation (R634H) in peripheral T-cell lymphoma, in which the arginine residues of STK10 were replaced with Histidines, with the subsequent decline in the pro-apoptotic activity of STK10. Also, other somatic mutations in STK10 have been reported, suggesting that STK10 may act as a suppressor gene (20).
We have experimentally demonstrated that STK10 plays a vital role in tumor cell migration and invasion. To reveal the underlying mechanism, we determined the phosphorylation and expression levels of ERM proteins, which have been demonstrated to be essential in the metastasis of various cancers (10,21-23). However, neither phosphorylation nor the expression level of all three ERM proteins is affected by the target deletion of STK10. Thus, STK10 participates in the biology of cervical cancer independently of the ERM pathway. To uncover the underlying mechanism, in the present study, we conducted an RNA-seq analysis for HeLa cells with and without STK10 and found the expression of 374 genes was changed in the cells lacking STK10.
Most importantly, more than half of these genes are related to tumor cell migration or invasion. PCDHG is a member of the protocadherin family and plays a vital role in mediating cell adhesion and synaptic development (24). FOXF2 and CDKN2A expression can significantly inhibit tumor cell migration and invasive ability (25,26). The downregulation of CTNNB1 can reduce the capacity for intercellular adhesion and promote cell migration and invasion (27). Compared with the control group, the expression levels of MUC1 in the STK10 KO HeLa and Caski cells were increased. MUC1 (Mucin-1) is a member of the mucoprotein family; the expression levels of MUC1 are upregulated in a variety of metastatic tumors (28-30). STK10 is an upstream kinase, acting as MAP kinase kinase kinase kinases (MAP4Ks). Maybe MUC1 is not the substrate of STK10 directly. However, we can conclude that the expression of MUC1 increases after STK10 depletion in cervical cancer cells, and the expression levels of other migration-related genes are also changed, jointly promoting the adhesion, migration, and invasion of cervical cancer cells, in a pathway that is ERM-independent.
Although more precise studies are needed to clarify the functional role of STK10 in the pathogenesis of cervical cancer, the current study has demonstrated for the first time a possible role of STK10 in the metastasis of cervical cancer and provided some new, useful information on the biology underlying cervical cancer, which will be of great benefit to patients through improved prevention, diagnosis, and treatment of this disease.
AcknowledgmentsOther Section
Funding: This work is supported by grants from
FootnoteOther Section
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1601
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1601
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1601). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Pimple S, Mishra G, Shastri S. Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol 2016;28:4-10. [Crossref] [PubMed]
- Denny L, de Sanjose S, Mutebi M, et al. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet 2017;389:861-70. [Crossref] [PubMed]
- Rauh-Hain JA, Clemmer JT, Bradford LS, et al. Racial disparities in cervical cancer survival over time. Cancer 2013;119:3644-52. [Crossref] [PubMed]
- Waggoner SE. Cervical cancer. Lancet 2003;361:2217-25. [Crossref] [PubMed]
- Leroy C, Belkina NV, Long T, et al. Caspase Cleavages of the Lymphocyte-oriented Kinase Prevent Ezrin, Radixin, and Moesin Phosphorylation during Apoptosis. J Biol Chem 2016;291:10148-61. [Crossref] [PubMed]
- Belkina NV, Liu Y, Hao JJ, et al. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc Natl Acad Sci U S A 2009;106:4707-12. [Crossref] [PubMed]
- McClatchey AI. ERM proteins. Curr Biol 2012;22:R784-5. [Crossref] [PubMed]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 2010;11:276-87. [Crossref] [PubMed]
- Louvet-Vallée S. ERM proteins: from cellular architecture to cell signaling. Biol Cell 2000;92:305-16. [Crossref] [PubMed]
- Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci 2015;128:1253. [Crossref] [PubMed]
- Hunter KW. Ezrin, a key component in tumor metastasis. Trends Mol Med 2004;10:201-4. [Crossref] [PubMed]
- Seton-Rogers S. Polarity: Merlin and ezrin get organized. Nat Rev Cancer 2013;13:76. [Crossref] [PubMed]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-11. [Crossref] [PubMed]
- Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46-53. [Crossref] [PubMed]
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-22. [Crossref] [PubMed]
- Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J BUON 2016;21:320-5. [PubMed]
- Wentzensen N, Arbyn M. HPV-based cervical cancer screening- facts, fiction, and misperceptions. Prev Med 2017;98:33-5. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Endo J, Toyama-Sorimachi N, Taya C, et al. Deficiency of a STE20/PAK family kinase LOK leads to the acceleration of LFA-1 clustering and cell adhesion of activated lymphocytes. FEBS Lett 2000;468:234-8. [Crossref] [PubMed]
- Fukumura K, Yamashita Y, Kawazu M, et al. STK10 missense mutations associated with anti-apoptotic function. Oncol Rep 2013;30:1542-8. [Crossref] [PubMed]
- Arpin M, Chirivino D, Naba A, et al. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr 2011;5:199-206. [Crossref] [PubMed]
- Gloerich M, Ponsioen B, Vliem MJ, et al. Spatial regulation of cyclic AMP-Epac1 signaling in cell adhesion by ERM proteins. Mol Cell Biol 2010;30:5421-31. [Crossref] [PubMed]
- Lallemand D, Arpin M. Moesin/ezrin: a specific role in cell metastasis? Pigment Cell Melanoma Res 2010;23:6-7. [Crossref] [PubMed]
- Yagi T. Clustered protocadherin family. Dev Growth Differ 2008;50:S131-40. [Crossref] [PubMed]
- Higashimori A, Dong Y, Zhang Y, et al. Forkhead Box F2 Suppresses Gastric Cancer through a Novel FOXF2-IRF2BPL-β-Catenin Signaling Axis. Cancer Res 2018;78:1643-56. [Crossref] [PubMed]
- Bai M, Yu NZ, Long F, et al. Effects of CDKN2A (p16INK4A/p14ARF) Over-Expression on Proliferation and Migration of Human Melanoma A375 Cells. Cell Physiol Biochem 2016;40:1367-76. [Crossref] [PubMed]
- Tian X, Liu Z, Niu B, et al. E-cadherin/β-catenin complex and the epithelial barrier. J Biomed Biotechnol 2011;2011:567305. [Crossref] [PubMed]
- Genitsch V, Zlobec I, Thalmann GN, et al. MUC1 is upregulated in advanced prostate cancer and is an independent prognostic factor. Prostate Cancer and Prostatic Diseases 2016;19:242-7. [Crossref] [PubMed]
- Ye Q, Yan Z, Liao X, et al. MUC1 induces metastasis in esophageal squamous cell carcinoma by upregulating matrix metalloproteinase 13. Lab Invest 2011;91:778-87. [Crossref] [PubMed]
- Shi M, Chen D, Yang D, et al. CCL21-CCR7 promotes the lymph node metastasis of esophageal squamous cell carcinoma by up-regulating MUC1. J Exp Clin Cancer Res 2015;34:149. [Crossref] [PubMed]


