
MiR-185-3p regulates epithelial mesenchymal transition via PI3K/Akt signaling pathway by targeting cathepsin D in gastric cancer cells
Introduction
Ranking as the fourth common cancer all around the world, gastric cancer (GC) remains the second cause for death among all the cancer-related death worldwide (1,2). Clinically, surgery with or without chemo or radio therapy is the most recommended curative approach. However, the poor prognosis, relatively short survival and rather low overall survival rate in advanced GC still raises the necessity for novel treatment methods with better therapeutic outcomes (2). It was well reported that the intratumor heterogeneity contributes dominantly to the malignancy of GC, emphasizing the importance to understand the mechanisms involved in the genetic alterations during the development and progression of GC.
Epithelial-mesenchymal transition (EMT) is a process during which the epithelial cells lose intercellular adhesion while gaining mesenchymal-like properties including increased invasion and migration capacity. EMT has been proved to participate in the occurrence and progress of cancers (3-5). It has been well established that EMT played a critical role in the formation, migration, invasion, and metastasis of GC, and that inhibition towards EMT could attenuate the progress of advanced GC (4-6). Therefore, it is necessary to explore the mechanism of EMT regulation within GC, which might alter current understanding towards the development and progress of GC, shedding light on novel therapeutic method to GC.
MicroRNAs (miRNAs) are a kind of conserved non-coding RNAs, with a length of 19 to 25 nucleotides (7). It has been well elucidated that miRNA could serve as either oncogene or tumor suppressor, making it a critical role in the comprehension toward tumor molecular pathogenesis (7,8). The downregulation of miR-125a-5p could promote cell proliferation in GC by boosting the expression of FOS1 (9), while miR-711 has been reported to suppress EMT in GC by downregulating the expression of CD44 (6). Besides, recently research reported that miR-185-3p could serve as an independent prognosis factor in GC, yet whether miR-185-3p could take a part in the regulation of EMT, as well as the detailed mechanism involved still remained unclear.
In the current research, we first proved that the miR-185-3p was commonly down-regulated in GCs, and that overexpression of miR-185-3p led to more apoptosis in GC cells. We then established a targeting relationship between miR-185-3p and cathepsin D. We also proved that the inhibition of miR-185-3p towards cathepsin D could affect the activity of PI3K/Akt signaling pathway, which further contributed to the regulation of EMT in GC cells. In addition, we found that overexpression of miR-185-3p led to the augmentation of apoptosis in GC cells. Our findings systematically demonstrated the role of miR-185-3p in the development and progression of GCs, which legitimated the potential of miR-185-3p as a novel therapeutic target for the treatment of GCs, shedding light on novel perspectives in the treatment towards GC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2133).
Methods
Patients and tissue specimens
Tumor and paired adjacent normal tissues were collected from 10 GC patients in Brain Hospital of Hunan Province for the analysis of clinicopathological characteristics. The expression level of miR-185-3p and cathepsin D in the collected tissues was analyzed by qRT-PCR. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of People’s Hospital of Hunan Province {approval no. [2018]-01.1} and informed consent was taken from all the patients.
Cell culture and reagents
Human gastric cancer cell lines MKN-1, AGS (CRL-1739), SGC7901 (CRL-1459), NCI-N87 (CRL-5822), BGC823 (CRL-2765), as well as human gastric epithelial cell line GES-1 were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA), and cultured in DMEM (Gibco, Gaithersburg, MD, USA) supplemented with 400 ng/mL hydrocortisone, 10% FBS and 1% penicillin/streptomycin. Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2, and used for study or split when almost confluent using trypsin/EDTA medium.
Plasmids and transfection
MiR-185-3p mimics, miR-185-3p inhibitor, pcDNA3.1-cathepsin D (p-cathepsin D) and the respective negative control plasmids were obtained from the GeneCopoeia Company (Guangzhou, China). AGS and SGC7901 cells were seeded into 6-well plates respectively and incubated for 18 h before transfection. Plasmids were then transfected into cells with Lipofectamine 2000® (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. The sequences for plasmids were as followed: for miR-185-3p mimics, sense, 5'-AGGGGCUGGCUUUCCUCUGGUC-3', and antisense, 5'-CCAGAG GAAAGCCAGCCCCUUU-3'; for miR-185-3p inhibitor, 5'-GACCAGAGGAAAGCCAGCCCCU-3'; for p-cathepsin D, sense, 5'-GGCTCCTCCAACCTGTGGGT-3', and antisense, 5'-CAGGTAGAAGGAGAAGATGT-3'; for NC mimics, sense, 5'-UUCUCCGAACGUGUCACGUTT-3', and antisense, 5'-ACGUGACACGUUCGGAGAATT-3'; for NC inhibitor, 5'-CAGUACUUUUGUGUAGUACAA-3'. The transfection efficiency was determined by fluorescent microscopy using fluorescein-labelled genes, and the expression level was evaluated by quantitative reverse transfection polymerase chain reaction. Transfected cells were collected after 48 h for analysis.
RNA extraction and quantitative reverse transfection polymerase chain reaction (qRT-PCR)
The expression level of miR-185-3p and cathepsin D in untreated GC cells, as well as the expression level of miR-185-3p, cathepsin D, and EMT markers (E-cadherin, vimentin, and N-cadherin) in cells treated with miR-185-3p mimics or miR185-3p inhibitor was evaluated by qRT-PCR. Total RNAs were extracted with TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA samples were reversed transcribed into cDNA with TaqMan Reverse Transcription Reagents (Life Technologies). The primers used in the study were as follows: for miR-185-3p, forward primer: 5'-ACACTCCAGCTGGGTGGAGAGAAAGGCAGT-3', and reverser primer: 5'-ACTGACTGATGCAATCTCAACTGGTGTCGTGGA-3'; for cathepsin D, forward primer: 5'-GTACATGATCCCCTGTGAGAAGGT-3', and reverse primer: 5'-GGGACAGCTTGTAGCCTTTGC-3'; for E-cadherin, forward primer: 5'-GGTTATTCCTCCCATCAGCT-3', and reverse primer 5'-CTTGGCTGAGGATGGTGTA-3'; for vimentin, forward primer: 5'-TGTCCAAATCGATGTGGATGTTTC-3', and reverse primer: 5'-TTGTACCATTCTTCTGCCTCCTG-3'; for N-cadherin, forward primer: 5'-GGTGGAGGAGAAGAAGACCAG-3', and reverse primer: 5'-GGCATCAGGCTCCACAGT-3'; for GAPDH, forward primer: 5'-GATTCCACCCATGGCAAATTC-3', and reverse primer: 5'-AGCATCGCCCCACTTGATT-3'. The quantification of miRNA PCR was calculated via 2−ΔΔCT method and normalized against GAPDH.
Western blotting analysis
The expression level of cathepsin D, E-cadherin, vimentin, N-cadherin, and the effectors in PI3K/Akt signaling pathway in the investigated cells was assessed by western blotting analysis. Briefly, cells were seeded into 6-well plates and incubated for 18 h before the assay. For the analysis toward cells transfected with miR-185-3p mimics, miR-185-3p inhibitor, NC mimics, or NC inhibitor, the assay started 72 h after the transfection. The cells were then harvested and lysed with Total Protein Extraction Kit (KeyGEN, Nanjing) according to the manufacture’s protocol, and 30 µg of the extracted protein was loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 4% stacking gel and 12% separating gel), in parallel with a molecular weight marker. The protein extracts were stacked at 80 V for 0.5 h and separated at 100 V for 2 h, thereafter, the samples were transferred to a nitrocellulose (NC) membrane and then blocked with 5% skim milk for 1.5 h at room temperature. Afterwards, the samples were incubated with primary antibodies at 4 °C overnight, which was followed by incubation with horseradish peroxidase-conjugated secondary antibody (1:1,000) at room temperature for 1.5 h. Finally, labelled bands from washed blots were detected by electrogenerated chemiluminescence (ECL) according to the manufacturer’s instructions. The quantification of the investigated protein expression level was normalized against GAPDH. Primary antibodies including cathepsin D (ab75852), E-cadherin (ab40772), N-cadherin (ab18203), vimentin (ab92547), PI3K (ab86714), p-PI3K (ab182651), Akt (ab8805), p-Akt (Ser 473) (ab8932) and GAPDH (ab8245) were purchased from Abcam (Cambridge, MA, USA).
Scratch assay
For cell scratch assay, cells were seeded into 6-well plates respectively and incubated for 18 h. Cells were then transfected with miR185-3p mimics, NC mimics, miR-185-3p inhibitor, or NC inhibitor. A 10 µL sterile pipette tip was used to scratch on the cell layer, and the gap closure was observed and recorded under microscope during the following 24 h.
Transwell migration assay
Cells were suspended in 200 µL serum-free medium and seeded into the upper chamber in Transwell inserts (Corning, Corning, NY, USA) with a membrane containing 8-mm pores. The inserts were then put into 24-well plate filled with conditioned medium, which served as the bottom chamber. Cells were incubated for 24 h, after which the cells on the filter surface were fixed with methanol, and stained using 0.1% crystal violet. The amounts of the migration cells in the lower chamber were observed and recorded under microscope.
Luciferase reporter construction and dual luciferase assay
Luciferase assay was performed to validate the targeting relationship between miR-185-3p and cathepsin D. Based on the results of the target prediction, where the 3' untranslated region (3'-UTR) sequence of cathepsin D contains the binding region with miR-185-3p, the luciferase reporter plasmids were obtained by inserting the 3'-UTR sequence of cathepsin D or a mutant sequence of cathepsin D into pGL3 promoter vector (Invitrogen), defining as pGL3-Cathepsin D-WT or pGL3-Cathepsin D-MUT respectively. AGS cells were seeded into 24-well plates and incubated for 18 h before the assay. Cells were first transfected with either pGL3-Cathepsin D-WT or pGL3-Cathepsin D-MUT, and then the cells were transfected with miR-185-3p mimics, NC mimics, miR-185-3p inhibitor, or NC inhibitor using Lipofectamine 2000® (Invitrogen Corp, CA, USA) according to the manufacturer’s introduction. The luciferase activity after transfection for 48 h was measured by the dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
Cell cycle and apoptosis evaluation
For cell cycle study, AGS and SGC7901 cells were seeded into 6-well plates respectively and incubated for 18 h. The cells were then transfected with miR185-3p mimics, NC mimics, miR-185-3p inhibitor, or NC inhibitor. Seventy-two hours after the transfection, the cells were harvested and fixed in 1 mL cold 70% ethanol for 24 h. Subsequently, the cells were centrifuged, washed, and incubated with 0.1% RNase A at 37 °C for 1 h. The cells were then stained with propidium iodide (PI) for 30 min at 4 °C and analyzed with flow cytometry. Cells treated with blank medium were used as the negative control.
Annexin V-PI (Beyotime Co., Ltd., Haimen, China) assay was used in the assessment of cell apoptosis. AGS and SGC7901 cells were first seeded into 6-well plates respectively and incubated for 18 h, then the cells were transfected with miR-185-3p mimics, NC mimics, miR-185-3p inhibitor, or NC inhibitor for 72 h. The cells were subsequently harvested and centrifuged, followed by incubation with 5 µL of Annexin V-FITC and 10 µL of PI solution for 15 min. The cells were analyzed using FACScan flow cytometry. Cells that were not treated with Annexin V-PI were used as controls.
Statistical analysis
All experiments were carried out triplicate. Data were presented as mean ± standard deviation (mean ± SD). A one-way analysis of variance (ANOVA) followed by Tukey post-hoc test or the Student’s t-test was used to compare the difference among various groups. Differences were considered statistically significant when P<0.05.
Results
MiR-185-3p was down-regulated in GC cells
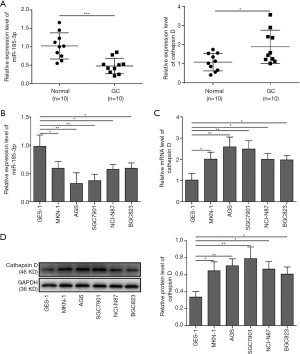
To explore whether miR-185-3p would affect the development of GCs, the expression level of miR-185-3p in 10 GC samples and paired adjacent normal tissue was evaluated with qRT-PCR. As shown in Figure 1A, the GC tissue exhibited lower expression of miR-185-3p compared with adjacent normal tissue, while the expression of cathepsin D was greatly elevated in tumor tissues, indicating a potential negative correlation between miR-185-3p and cathepsin D which might interfere with the development of GCs. For an accurate assessment of the correlation among miR-185-3p, cathepsin D, and tumor progression, we further determined the expression level of miR-185-3p and cathepsin D in various GC cell lines via qRT-PCR and Western blotting analysis. As suggested in Figure 1B, compared with gastric epithelial cells (GES-1), all the tested GC cell lines showed a lower expression level of miR-185-3p. On the contrary, the mRNA level of cathepsin D in the tested GC cell lines was all higher than that in GES-1 (Figure 1C). The results of Western blotting (Figure 1D) also validated this finding, where the expression level of cathepsin D in GES-1 was obviously lower than its expression in GC cells. These results indicated that there might be a correlation between the down-regulated miR-185-3p and the increased expression of cathepsin D. Based on the findings, AGS and SGC7901 were selected in the following study, for these 2 cell lines showed the lowest level of miR-185-3p.
Overexpression of miR-185-3p inhibited EMT, led to cell cycle arrest and induced apoptosis in GC cell lines
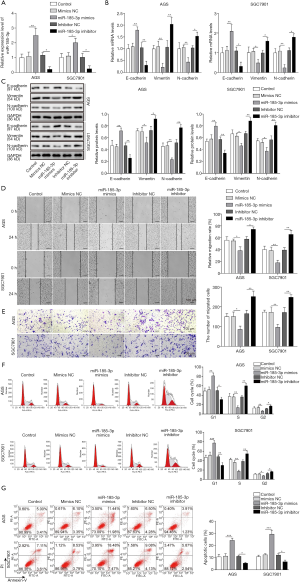
The inhibition of miR-185-3p towards EMT in AGS and SGC7901 cells was confirmed on both molecular level and cellular level. The expression level of EMT markers, including epithelial marker E-cadherin, as well as mesenchymal markers vimentin and N-cadherin was first evaluated via qRT-PCR and Western blotting analysis. The successful transfection of miR-185-3p in both investigated cell lines (Figure 2A) led to a drastic decrease in the expression level of mesenchymal markers, while the expression level of epithelial marker was noticeably elevated by the boosted expression of miR-185-3p. On the contrary, knockdown of miR-185-3p resulted in an increased mRNA and protein level of mesenchymal markers and a reduced expression of epithelial marker (Figure 2B,C). The influence of miR-185-3p on EMT markers was in consistency with its inhibition towards cell migration. As the result of cell scratch assay suggested, the overexpression of miR-185-3p inhibited the migration capacity of both AGS cells and SGC 7901 cells (Figure 2D), while knockdown of miR-185-3p resulted in a promotion of migration capacity. The results of Transwell assay further validated the influence of miR-185-3p on the cell migration ability, where the upregulation of miR-185-3p led to a compromised migration capacity of AGS cells and SGC 7901 (Figure 2E) and knockdown of miR-185-3p resulted in a promotion of migration capacity. Besides, miR-185-3p also exerted an influence on the cell cycle distribution. As suggested in Figure 2F, overexpression of miR-185-3p contributed to an elevated ratio of AGS cells blocked in G0/G1 phase. The cell cycle arrest in G0/G1 phase consequently led to augmented apoptosis in AGS and SGC7901 cells. However, knockdown of miR-185-3p showed the opposite phenomenon (Figure 2G). All above data indicated that overexpression of miR-185-3p inhibited the proliferation, EMT, and migration of GC cells, while the inhibition of miR-185-3p showed the opposite effect.
MiR-185-3p targeting inhibited the expression of cathepsin D in GC cells
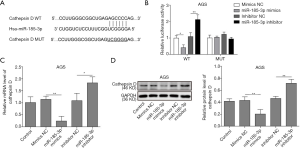
MiR-185-3p could bind with cathepsin D gene on the 3'-UTR region according to the results bioinformation analysis (Figure 3A), based on which a dual luciferase assay was performed to investigate the targeting relationship between miR185-3p and cathepsin D. As shown in Figure 3B, overexpression of miR-185-3p remarkably reduced the luciferase activity in wild type AGS cells (AGS-WT), while the inhibition of miR-185-3p contributed to an enhanced luciferase activity in AGS-WT cells. However, neither overexpression nor inhibition of miR-185-3p affected the luciferase activity in mutated AGS cells (AGS-MUT), validating the result in Figure 3A, where miR-185-3p could bind with cathepsin D gene directly. The influence of miR-185-3p on the expression level of cathepsin D was also investigated to further confirm the predicted targeting relationship. Both qRT-PCR and Western blotting analysis showed that overexpressed miR-185-3p led to a decrease expression of cathepsin D, while that the inhibition towards miR-185-3p resulted in a boosted level of cathepsin D (Figure 3C,D). Therefore, it is reasonable to draw a conclusion that a targeting inhibition relationship existed between miR-185-3p and cathepsin D.
MiR-185-3p inhibited PI3K/Akt axis via regulating cathepsin D
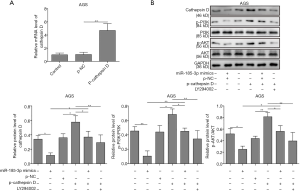
Overexpressed miR-185-3p has been reported to inhibit the activity of PI3K/Akt signaling pathway (10,11). To explore whether cathepsin D was involved in the above-mentioned inhibition, the expression level of PI3K, phosphorylated PI3K (p-PI3K), AKT, and the phosphorylation of AKT on Ser 473 site [p-AKT (Ser 473)] was determined in AGS cells. p-cathepsin D was transfected into AGS cells, which resulted in a robustly high level of cathepsin D (Figure 4A). As results in Figure 4B suggested, the overexpression of miR-185-3p inhibited the activity of PI3K/Akt axis, while the overexpression of cathepsin D contributed to the activation of PI3K/Akt signaling pathway, The activation led by cathepsin D could be neutralized or even reversed by the overexpression of miR-185-3p or the existence of PI3K/Akt inhibitor (LY294002), where a remarkably decreased level of cathepsin D could also be observed. Therefore, it was clear that a positive regulation relationship existed between cathepsin D and the activity of PI3K/Akt signaling pathway, and that miR-185-3p could inhibit PI3K/Akt axis by targeting cathepsin D.
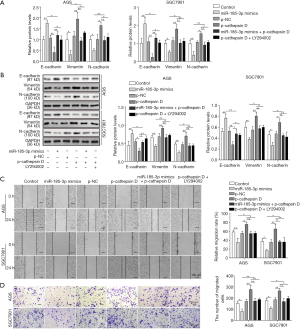
MiR-185-3p regulated EMT via cathepsin D mediated PI3K/Akt signaling pathway
As one of the most frequently dysregulated intracellular signaling pathway, PI3K/Akt was reported to play an important part in EMT (3,12-14). Therefore, we studied the role of miR-185-3p in the regulation of EMT. As shown in Figure 5A,B, the expression level of E-cadherin was reduced by the overexpression of cathepsin D, while the level of both vimentin and N-cadherin was elevated, suggesting that cathepsin D might promote the malignancy of GC by facilitating EMT. Yet it was noticeable that the influence of overexpressed cathepsin D towards all the EMT-related proteins was compromised by either miR-185-3p or LY294002, indicating that interference in PI3K/Akt signaling pathway could affect the promotion towards cell migration capacity brought by cathepsin D. This result was in consistency with cell migration studies. As shown in Figure 5C, the migration capacity of AGS cells was enhanced with boosted level of cathepsin D, which could be ameliorated by the existence of either overexpressed miR-185-3p or LY294002. The results of Transwell assay further validated this finding, where the migration ability of AGS cells was significantly facilitated by increased level of cathepsin D, and then attenuated by miR-185-3p or PI3K/Akt inhibitor (Figure 5D). Comprehensively, all these data revealed that the regulation of miR-185-3p on EMT was based on its inhibition toward cathepsin D, and that by targeting cathepsin D, miR-185-3p could regulate the activity of PI3K/Akt signaling pathway, resulting in its function towards EMT progression.
Discussion
The complicity and high heterogeneity in GC have long been proved to contribute to the failure in the treatment of GC, which remains a major clinical hindrance (1,15), calling for a deeper understanding for an altered therapy for GC. In our current research, we innovatively demonstrated a targeting relationship between miR-185-3p, a highly inhibited miRNA in GC, and cathepsin D, through which the activity of PI3K/Akt signaling pathway would be affected, consequently leading to a facilitated malignancy of GC cells. The finding of this study broadened our comprehension of the genetic alterations in GC cells, which may pave the way to the development of gene therapy towards GC.
Inspired by previous studies, we found that miR-185-3p was generally down-regulated in GC tissues collected from patients, as well as in various GC cell lines (16-18). It was also well established that miR-185-3p could serve as a potential therapeutic target for multiple cancers, the boosting of which could led to the suppression towards tumor development, metastasis, as well as chemo- and radio-resistance (19-24). Therefore, the dysregulated level of miR-185-3p in GC cells indicated that it may also play a dominant part in the progression of GC.
By elevating the expression level of miR-185-3p in GC cells, we found that the EMT progress as well as cancer migration capacity was compromised. This finding was in consistency with former studies, where the tumor suppression effect of miR-185-3p was attributed to its inhibition toward EMT by interacting with the 3'-UTR of certain genes, including PTPN-14, STIM1, and so on (22,25-27). Yet it was noteworthy that our findings innovatively confirmed the direct inhibition of miR-185-3p on cathepsin D, which may provide a precise and clear explanation of the suppression towards EMT induced by overexpressed miR-185-3p. cathepsin D, a frequently overexpressed protein in various cancers, was well proved to promote EMT in GC by interfering with apoptosis in cancer cells (28,29). In present study, by evaluating the influence of overexpressed miR-185-3p on cell EMT progress and migration capacity, we proved that the dynamics between miR-185-3p and cathepsin D participated in the regulation of EMT and migration in GC cells, which would lead to the interference with the progression of GC as a result.
Apart from regulating the expression level of several critical proteins in tumor development, emerging evidence also elucidated that miR-185-3p interferes with tumor progression by regulating PI3K/Akt axis (10,22,24,30,31). In the current research, the activity of PI3K/Akt signaling pathway was significantly enhanced with an increased level of cathepsin D, but obviously compromised after inhibiting cathepsin D by overexpressed miR-185-3p. The same trend also went to the migration capacity of cells, where the promoted migration capacity of cells by elevation in cathepsin D was ameliorated by the overexpression of miR-185-3p. In addition, a promotion in apoptosis was observed in GC cells with overexpressed miR-185-3p, further validating that miR-185-3p could serve as a novel therapeutic target for the treatment of GCs. These findings indicated that by targeting inhibition towards cathepsin D, miR-185-3p could regulate the activity of PI3K/Akt signaling pathway, consequently inhibiting the proliferation, compromising EMT process, and limiting the migration of cancer cells, which would finally attenuate the malignancy of GC.
Conclusions
In a summary, we demonstrated that expression of miR-185-3p was decreased and cathepsin D was increased in GC cells. MiR-185-3p inhibited EMT by targeting cathepsin D in GC cell lines, and PI3K/Akt signaling pathway involved in it. Our findings suggested that boosting the expression of miR-185-3p or targeting inhibition towards cathepsin D could serve as a potential therapy for GCs. Yet this research mainly focused on the in vitro study, therefore, we would further explore the targeting relationship between miR-185-3p and cathepsin D in vivo as well as in clinical, in the hope of establishing a novel treatment for GCs with a facilitated therapeutic outcome.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2133
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2133
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (Available at http://dx.doi.org/10.21037/tcr-19-2133). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of People’s Hospital of Hunan Province {approval no. [2018]-01.1} and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Sitarz R, Skierucha M, Mielko J, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res 2018;10:239-48. [Crossref] [PubMed]
- Wang R, Song Y, Liu X, et al. UBE2C induces EMT through Wnt/beta-catenin and PI3K/Akt signaling pathways by regulating phosphorylation levels of Aurora-A. Int J Oncol 2017;50:1116-26. [Crossref] [PubMed]
- Xia P, Xu XY. Epithelial-mesenchymal transition and gastric cancer stem cell. Tumour Biol 2017;39:1010428317698373. [Crossref] [PubMed]
- Xu GF, Zhang WJ, Sun Q, et al. Combined epithelial-mesenchymal transition with cancer stem cell-like marker as predictors of recurrence after radical resection for gastric cancer. World J Surg Oncol 2014;12:368. [Crossref] [PubMed]
- Xiao WS, Li DF, Tang YP, et al. Inhibition of epithelial-mesenchymal transition in gastric cancer cells by miR-711-mediated downregulation of CD44 expression. Oncol Rep 2018;40:2844-53. [Crossref] [PubMed]
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [Crossref] [PubMed]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019;20:5-20. [Crossref] [PubMed]
- Wang S, Ran LK, Zhang WF, et al. FOXS1 is regulated by GLI1 and miR-125a-5p and promotes cell proliferation and EMT in gastric cancer. Sci Rep-Uk 2019; 9.
- Zhang S, Li D, Jiao G J, et al. miR-185 suppresses progression of Ewing's sarcoma via inhibiting the PI3K/AKT and Wnt/beta-catenin pathways. Onco Targets Ther 2018;11:7967-77. [Crossref] [PubMed]
- Tsitoura E, Wells AU, Karagiannis K, et al. MiR-185/AKT and miR-29a/collagen 1a pathways are activated in IPF BAL cells. Oncotarget 2016;7:74569-81. [Crossref] [PubMed]
- Cheng N, Li HX, Luo JP. Trop2 promotes proliferation, invasion and EMT of nasopharyngeal carcinoma cells through the NF-kappa B pathway. RSC Advances 2017;7:53087-96. [Crossref]
- Song Y, Li ZX, Liu X, et al. The Wnt/β-catenin and PI3K/Akt signaling pathways promote EMT in gastric cancer by epigenetic regulation via H3 lysine 27 acetylation. Tumour Biol 2017;39:1010428317712617. [Crossref] [PubMed]
- Silva EM, Begnami MD, Fregnani JH, et al. Wnt and AKT/PI3K Signaling Pathway Markers Pattern Expression and Its Association with Clinicopathological Features and Prognosis in Gastric Tumors. Gastroenterology 2009;136:A747. [Crossref]
- Tran P, Nguyen C, Klempner SJ. Targeting the Phosphatidylinositol-3-kinase Pathway in Gastric Cancer: Can Omics Improve Outcomes? Int Neurourol J 2016;20:S131-S140. [Crossref] [PubMed]
- Xiang Y, Ma N, Wang D, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene 2014;33:378-86. [Crossref] [PubMed]
- Zhi Q, Zhu J, Guo X, et al. Metastasis-related miR-185 is a potential prognostic biomarker for hepatocellular carcinoma in early stage. Biomed Pharmacother 2013;67:393-8. [Crossref] [PubMed]
- Qu F, Cui X, Hong Y, et al. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol Cell Biochem 2013;377:121-30. [Crossref] [PubMed]
- Imam JS, Buddavarapu K, Lee-Chang JS, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 2010;29:4971-9. [Crossref] [PubMed]
- Liao JM, Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. J Biol Chem 2011;286:33901-9. [Crossref] [PubMed]
- Ma D, Cao Y, Wang Z, et al. CCAT1 lncRNA Promotes Inflammatory Bowel Disease Malignancy by Destroying Intestinal Barrier via Downregulating miR-185-3p. Inflamm Bowel Dis 2019;25:862-74. [Crossref] [PubMed]
- Liu C, Li G, Ren S, et al. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett 2017;13:2631-6. [Crossref] [PubMed]
- Li Q, Wang JX, He YQ, et al. MicroRNA-185 regulates chemotherapeutic sensitivity in gastric cancer by targeting apoptosis repressor with caspase recruitment domain. Cell Death Dis 2014;5:e1197. [Crossref] [PubMed]
- Zhao L, Zhang Y, Liu J, et al. miR-185 Inhibits the Proliferation and Invasion of Non-Small Cell Lung Cancer by Targeting KLF7. Oncol Res 2019;27:1015-23. [Crossref] [PubMed]
- Zhang Z, Liu X, Feng B, et al. STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene 2015;34:4808-20. [Crossref] [PubMed]
- Tan Z, Jiang H, Wu Y, et al. miR-185 is an independent prognosis factor and suppresses tumor metastasis in gastric cancer. Mol Cell Biochem 2014;386:223-31. [Crossref] [PubMed]
- Yin C, Zhang G, Sun R, et al. miR-185-5p inhibits F-actin polymerization and reverses epithelial mesenchymal transition of human breast cancer cells by modulating RAGE. Mol Med Rep 2018;18:2621-30. [Crossref] [PubMed]
- Ishimoto T, Baba H, Izumi D, et al. Current perspectives toward the identification of key players in gastric cancer microRNA dysregulation. Int J Cancer 2016;138:1337-49. [Crossref] [PubMed]
- Zhang M, Wu JS, Yang X, et al. Overexpression Cathepsin D Contributes to Perineural Invasion of Salivary Adenoid Cystic Carcinoma. Front Oncol 2018;8:492. [Crossref] [PubMed]
- Zhou L, Liu S, Han M, et al. miR-185 Inhibits Fibrogenic Activation of Hepatic Stellate Cells and Prevents Liver Fibrosis. Mol Ther Nucleic Acids 2018;10:91-102. [Crossref] [PubMed]
- Zhou CW, Zhao WJ, Zhu YG, et al. MiR-185 inhibits tumor growth and enhances chemo-resistance via targeting SRY-related high mobility group box transcription factor 13 in non-small-cell carcinoma. Am J Transl Res 2018;10:2600-9. [PubMed]


