
Bioinformatics analysis of microenvironment-related genes associated with radioresistance in glioblastoma
Introduction
Glioblastoma (GBM) is the most common primary brain tumor in adults, and has a high rate of mortality. Despite improvements in therapeutic strategies, its clinical outcomes remain poor, with a median survival time of approximately 15 months (1). GBM cells exhibit aggressive behavior, with high levels of invasion and diffuse infiltration into the surrounding brain parenchyma, which makes complete tumor resection impossible (2). Radiotherapy (RT) is an effective treatment modality for eliminating residual tumor cells. Unfortunately, most patients with GBM experience local recurrence at the original lesion sites where high doses of radiation were delivered, confirming the radioresistance of GBM (3). To further improve the efficacy of RT, the underlying mechanism of GBM radioresistance should be determined, and biomarkers for assessing treatment responses and prognosis must be identified.
GBM cells coexist with non-tumor cells in a dynamic microenvironment, which promotes GBM proliferation, invasion, and survival. Numerous studies have shown that the intrinsic characteristics of GBM cells as well as their interactions with the microenvironment contribute towards GBM resistance to RT (4). The GBM microenvironment contains a diverse array of non-tumor cells, including immune cells, stromal cells, and endothelial cells, as well as extracellular matrix components. The two major types of cells in the GBM microenvironment are infiltrating immune cells (such as microglia, macrophages, lymphocytes, neutrophils, and dendritic cells) and stromal cells (such as neurons, astrocytes, and oligodendroglia) (5). Several findings have demonstrated the critical roles of immune and stromal cells in the prognostic assessment of tumors.
Using public databases and novel biological algorithms that provide survival estimates for patients, considerable progress has been made in predicting cancer prognosis based on clinical features and gene-expression profiles (6). The recently developed Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm can be used to evaluate stromal and immune scores, and to predict the infiltration of non-tumor cells based on their unique gene-expression signatures (7). The ESTIMATE algorithm has been applied for predicting prognosis and for identifying genetic alterations in many cancers, including GBM (8-10). Although Jia et al. identified genes with prognostic value in the GBM microenvironment, the relationship between microenvironment-related genes and GBM radioresistance is unclear (8). Thus, immune and stromal scores can be used to assess the radioresponse of GBM from a genome-wide perspective.
Here, we evaluated the associations between immune and stromal scores, and the clinical outcomes of patients with GBM undergoing RT (GBM-RT), using data from The Cancer Genome Atlas (TCGA) database and the ESTIMATE algorithm. We identified microenvironment-related genes that correlated with radioresistance and a poor prognosis for GBM, which were validated using data from the Chinese Glioma Genome Atlas (CGGA) cohort. Understanding the relationship between the immune microenvironment and GBM radiosensitivity may aid the development of strategies to improve the efficacy of RT.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2476).
Methods
Databases
The RNA-sequencing (RNA-seq) data and clinical information for patients with primary GBM, including 348 patients who underwent RT treatment and had detailed survival-time records, were retrieved from TCGA (https://tcga-data.nci.nih.gov/tcga/). For the validation cohort, RNA-seq data and clinical information for patients with primary GBM, including 71 patients who underwent RT treatment and had detailed survival-time records, were extracted from the CGGA (http://www.cgga.org.cn/). The ESTIMATE algorithm was used to evaluate the proportion of non-tumor components in the tumor microenvironment according to their gene-expression signatures, which are represented by the ImmuneScore, StromalScore, and ESTIMATEScore (7). The ImmuneScore and StromalScore of TCGA GBM cases were available from a public source website (https://bioinformatics.mdanderson.org/estimate/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed genes (DEGs)
The limma package of R software was utilized to identify DEGs by comparing the high-score and low-score cases (11). Gene-expression levels with a |log2 (fold-change)| score of >1 and an adjusted (adj.) P value of <0.05 were considered significantly different.
Heatmaps, volcano plots, and Venn diagrams
Heatmaps of DEGs were constructed using the ClustVis web tool (https://biit.cs.ut.ee/clustvis/), and volcano plots for DEGs were generated with GraphPad Prism. DEGs common to different groups were identified by generating Venn diagrams, which were created using an online analysis tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) (12).
Enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
The clusterProfiler, org.Hs.eg.db, enrichplot, and ggplot2 packages of R software were used to perform GO and KEGG enrichment analysis of DEGs. GO categories were analyzed, including biological processes (BPs), molecular functions (MFs), and cellular components (CCs). Categories with P and q values of <0.05 were regarded as being significantly enriched.
Protein-protein interaction (PPI) network construction
The PPI network of the common DEGs was generated using the STRING database (http://string-db.org) (13). An interaction score of ≥0.4 was selected as the cutoff criterion. PPI network data were reconstructed using Cytoscape software to identify genes and to perform module analysis. Molecular Complex Detection was applied to detect the clusters in the network, and individual networks with more than 10 nodes were included.
Statistical analysis
Statistical analyses were conducted using R software (version 4.0.0) and GraphPad Prism (version 7.0.0; GraphPad, Inc., La Jolla, CA, USA). Data for 348 patients with primary GBM (TCGA GBM database) were assigned high or low immune and stromal scores relative to the median ImmuneScore and StromalScore, respectively. Individual DEGs were grouped as having high or low expression relative to the median score of each individual gene-expression value. The Kaplan-Meier method was used for survival analysis and P<0.05 (determined with the log-rank test) was regarded as reflecting a statistically significant difference. To determine the prognostic values of DEGs, the survival package of R software was used for univariate Cox regression analysis, and P<0.05 was considered a significant difference.
Workflow
The workflow of the current study is shown in Figure S1. RNA-seq data and clinical information for 539 patients were downloaded from TCGA database, of which 348 cases undergoing RT were included in this study. ESTIMATE scores, including stromal and immune scores, were obtained using the gene expression data from GBM tumor tissues. The impact of ESTIMATE scores on the prognosis of patients with GBM-RT was investigated. Shared DEGs were obtained by intersection analysis between the PPI network and univariate Cox regression analyses. DEGs with prognostic value for RT were further validated using the CGGA GBM database.
Results
Tumor stromal and immune scores correlated with the clinical outcomes of patients with GBM-RT
To investigate correlations between the overall survival (OS) of patients with GBM-RT and their stromal/immune scores, TCGA data from 348 patients with GBM-RT were obtained and divided into high and low groups (RT+ stromal scorehigh, RT+ stromal scorelow, RT+ immune scorehigh, and RT+ immune scorelow groups), based on their stromal and immune scores. The median values of the stromal and immune scores were 84.5 and 966.78, respectively. Survival curves showed that patients with high stromal scores had a shorter OS than those with low stromal scores (Figure 1A, P=0.0166; log-rank test) and that patients with high immune scores tended to have a shorter median OS (Figure 1B, P=0.0661; log-rank test). These results indicate that the stromal or immune score plays a role in determining the clinical outcomes of patients with GBM-RT. Moreover, the stromal score showed greater prognostic value than the immune score and was more suitable for stratifying the radioresponses of patients with GBM.

Associations between gene-expression profiles and stromal and immune scores
To determine the gene expression profiles, differential analysis of 348 patients with GBM-RT was performed according to the stromal or immune scores. DEGs were identified using a threshold of |log2 (fold-change)| >1 and an adj. P value of <0.05. We identified 168 and 205 DEGs that were upregulated in the high-stromal score and high-immune score groups, respectively (Figure 1C,D, Figure S2A,B). Venn diagrams were used to identify critical genes relevant to the GBM microenvironment; 139 upregulated DEGs overlapped with the high-stromal and high-immune score groups, whereas 13 downregulated DEGs were identified (Figure S2C,D).
Functional enrichment analysis of DEGs in patients with GBM-RT
To analyze the underlying biological functions of DEGs, functional enrichment clustering of the 139 shared DEGs was performed. We identified 359, 36, and 62 GO terms representing BPs, MFs, and CCs, respectively, that were significantly enriched (Figure 2A,B, Figure S3, and Tables S1-S3; q<0.05 and adj. P<0.05). The top GO terms included antigen processing and presentation, response to interferon-gamma, major histocompatibility complex (MHC) class II protein complex, peptide antigen binding, and immune receptor activity. KEGG analysis revealed enrichment of 56 pathways (q<0.05 and adj. P<0.05) involved in various aspects of inflammation and immunity, including Staphylococcus aureus infection, inflammatory bowel disease, the IL-17 signaling pathway, NF-κB signaling pathway, TNF signaling pathway, cytokine-cytokine receptor interaction, and Toll-like receptor signaling pathway (Figure 2C,D, Table S4). The results demonstrate that the biological functions of the DEGs were linked to inflammation or immune-related activities, indicating that inflammation or immune features in the GBM microenvironment mediate GBM radioresistance.

PPIs among genes with radioresistance signatures
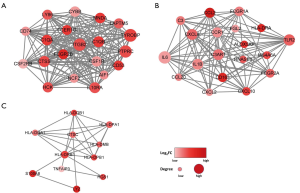
A PPI network was created to investigate the roles of the identified DEGs in GBM radioresistance. The network included 132 nodes and 1,357 edges, and consisted of three significant modules, based on the results of the Molecular Complex Detection method. The representative nodes were TYROBP (Figure 3A), IL-6 (Figure 3B), and HLA-DRB1 (Figure 3C), which were closely connected with other nodes in each module.
Identification of prognostic DEGs in patients with GBM-RT
Next, Cox regression analysis was conducted to investigate the roles of identified DEGs in the prognosis of patients with GBM-RT. A total of 88 DEGs (86 and 2 shared DEGs upregulated in the high- and low-expression stromal and immune groups, respectively) were shown to significantly correlate with the OS of patients with GBM-RT (Table S5; P<0.05). Furthermore, 29 DEGs were obtained by overlapping the top nodes in the PPI network, and 86 significant DEGs were identified by Cox regression. Among the 29 overlapping genes, log-rank tests and Kaplan-Meier plots indicated that 19 genes had prognostic value in patients with GBM-RT (Figure 4A,B,C,D and Figure S4) and were associated with a poor response to RT.

Validation of DEGs using the CGGA database
To determine whether the DEGs in TCGA database had prognostic significance in other cases of GBM, the 19 genes were further investigated using data from the CGGA database. The gene-expression data of 325 patients with glioma, including 71 with GBM-RT, were downloaded and analyzed. We found that 10 of the 19 genes were significantly linked to a poor prognosis in patients with GBM-RT in the CGGA cohort, according to the log-rank test. These genes included TLR2, C3AR1, CD163, ALOX5AP, NCF2, CYBB, FCGR1A, FCGR2A, FCGR2B, and RNASE6. The Kaplan-Meier plots of all 10 genes are shown in Figure 4E,F,G,H and Figure S5.
Discussion
Radioresistance is considered the main cause of early recurrence and unsatisfactory clinical outcomes in patients with GBM. Thus, exploring the molecular basis of radioresistance is essential for improving the efficacy of RT in GBM management. The interplay between RT and the GBM microenvironment is reported to contribute to radioresistance development (14). Of the various stromal cells in the microenvironment, astrocytes interact most frequently with GBM cells to mediate the radioresponse (15). With respect to immune cells, macrophages and microglia in the microenvironment induce stemness and chemo-radioresistance in GBM cells (16). Additionally, changes in the GBM microenvironment induced by RT can in turn contribute to radioresistance, leading to tumor relapse (17).
We identified the GBM microenvironment-related genes in TCGA database that appeared to contribute to GBM radioresistance, and verified the gene signature using the CGGA database. First, we determined the stromal and immune scores, and investigated their relationships with the prognoses of patients with GBM-RT. Notably, a previous report revealed no significant correlation between OS and the immune and stromal scores of patients with GBM (8). In contrast, our results indicated that higher stromal scores were significantly correlated with a shorter OS in patients with GBM-RT, suggesting that the stromal score is a negative prognostic indicator that can be used to predict the radioresponses of patients with GBM. Next, we established the global-expression profiles of genes in the GBM microenvironment, according to the stromal or immune scores. Functional enrichment analyses indicated that these genes participated in inflammation and immune activities, suggesting that immune features in the GBM microenvironment play vital roles in mediating GBM radioresistance. PPI-network analysis revealed that 19 DEGs had prognostic value and were associated with radioresistance. To confirm the validity and reliability of these results, these genes were further validated using data from the CGGA cohort. Finally, 10 DEGs were found to be significantly related to the poor prognosis of patients with GBM-RT and to radioresistance. Three of these DEGs including CD163, TLR2, and C3AR1 have been reported to play potential roles in the resistance of tumor cells to RT.
CD163 is a marker of tumor-associated macrophages. Increasing evidence has revealed that accumulation of CD163-positive tumor-associated macrophages plays important roles in tumor invasion, progression, and poor clinical outcomes in patients with breast cancer (18) and nasopharyngeal carcinoma (19). In the case of breast cancer, patients with CD163-positive tumors had a shorter OS following postoperative RT, suggesting that CD163 is involved in radioresistance. This possibility was confirmed by conducting in vitro experiments (20). Additionally, CD163 was also reported to be overexpressed in glioma cells and to contribute to an unfavorable prognosis in patients with GBM (21). Notably, CD163 is a specific marker of M2 macrophages in patients with GBM (22), and is known to be involved in GBM radioresistance (23). TLR2 is a member of the TLR family, which comprises critical modulators of the innate immune responses to pathogen- and damage-associated molecules. TLR2 overexpression is known to be related to increased risks of tumorigenesis and poor outcomes in patients with gastric cancer (24), prostate cancer (25), and colon cancer (26). In a murine model of orthotopic glioma, TLR2 activation of microglia promoted glioma immune-system evasion (27). Consistent with our results, Li et al. showed that TLR2 was positively associated with the glioma grade and poor OS, and that TLR2 overexpression contributed to glioma progression (28). Notably, radioresistant effects of TLR2 were also demonstrated. In vivo experiments showed that TLR2-knockout mice were more susceptible to irradiation-induced death, whereas treatment with the TLR2-ligand Pam3CSK4 induced radioresistance (29). However, TLR2 activation has also been reported to play a role in innate and adaptive immunity against brain tumors (30). Thus, the specific relationship between TLR2 and both GBM tumor progression and radioresistance requires further investigation. C3AR1 encodes the complement C3a receptor 1 (C3aR1), a complement cascade receptor that functions as an immune regulator. Findings with experimental tumor models have indicated that signaling through C3aR1 can impact tumor growth by modifying immune infiltrates in the tumor microenvironment (31). C3AR1 upregulation was recently reported to correlate with chemoresistance and the survival of patients with soft tissue sarcoma (32). Using weighted gene co-expression network analysis, Pan et al. also identified several immune inflammatory response-related genes (including C3AR1) in GBM, particularly in the mesenchymal subtype (33). It has been proposed that C3aR1 is upregulated in response to RT, which affects RT-induced tumor-specific immunity (34).
One limitation of our study was that our research was retrospective. Therefore, prospective studies are needed to confirm our results. In addition, the gene signatures were analyzed based on TCGA and CGGA data, and these findings should be verified in clinical, cellular, and animal experiments.
Conclusions
In conclusion, using the ESTIMATE algorithm, our bioinformatics analysis of patients with GBM-RT enabled the identification of GBM microenvironment-related genes associated with radioresistance. The stromal/immune score-based gene signatures identified here represent promising biomarkers for GBM and provide a theoretical basis for predicting the radioresponses and clinical outcomes of patients with GBM. However, further investigation is necessary to examine potential associations between these genes and RT combined with immunotherapy.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2476
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2476
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2476). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Urbańska K, Sokołowska J, Szmidt M, et al. Glioblastoma multiforme - an overview. Contemp Oncol (Pozn) 2014;18:307-12. [Crossref] [PubMed]
- Gupta K, Burns TC. Radiation-induced alterations in the recurrent glioblastoma microenvironment: therapeutic implications. Front Oncol 2018;8:503. [Crossref] [PubMed]
- Zhang H, Zhou Y, Cui B, et al. Novel insights into astrocyte-mediated signaling of proliferation, invasion and tumor immune microenvironment in glioblastoma. Biomed Pharmacother 2020;126:110086. [Crossref] [PubMed]
- Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell 2017;31:326-41. [Crossref] [PubMed]
- Zhu W, Xie L, Han J, et al. The application of deep learning in cancer prognosis prediction. Cancers (Basel) 2020;12:603. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Jia D, Li S, Li D, et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY) 2018;10:592-605. [Crossref] [PubMed]
- Bi KW, Wei XG, Qin XX, et al. BTK has potential to be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment remodeling: a study based on TCGA data mining. Front Oncol 2020;10:424. [Crossref] [PubMed]
- Wang H, Wu X, Chen Y. Stromal-immune score-based gene signature: a prognosis stratification tool in gastric cancer. Front Oncol 2019;9:1212. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Lin G, Chai J, Yuan S, C, et al. VennPainter: a tool for the comparison and identification of candidate genes based on Venn diagrams. PLoS One 2016;11:e0154315. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Stapleton S, Jaffray D, Milosevic M. Radiation effects on the tumor microenvironment: Implications for nanomedicine delivery. Adv Drug Deliv Rev 2017;109:119-30. [Crossref] [PubMed]
- Rath BH, Wahba A, Camphausen K, et al. Coculture with astrocytes reduces the radiosensitivity of glioblastoma stem‐like cells and identifies additional targets for radiosensitization. Cancer Med 2015;4:1705-16. [Crossref] [PubMed]
- Hide T, Shibahara I, Kumabe T. Novel concept of the border niche: glioblastoma cells use oligodendrocytes progenitor cells (GAOs) and microglia to acquire stem cell-like features. Brain Tumor Pathol 2019;36:63-73. [Crossref] [PubMed]
- Dahan P, Martinez Gala J, Delmas C, et al. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Cell Death Dis 2014;5:e1543. [Crossref] [PubMed]
- Ramos RN, Rodriguez C, Hubert M, et al. CD163+ tumor-associated macrophage accumulation in breast cancer patients reflects both local differentiation signals and systemic skewing of monocytes. Clin Transl Immunology 2020;9:e1108. [Crossref] [PubMed]
- Yu Y, Ke L, Lv X, et al. The prognostic significance of carcinoma-associated fibroblasts and tumor-associated macrophages in nasopharyngeal carcinoma. Cancer Manag Res 2018;10:1935-46. [Crossref] [PubMed]
- Garvin S, Oda H, Arnesson LG, et al. Tumor cell expression of CD163 is associated to postoperative radiotherapy and poor prognosis in patients with breast cancer treated with breast-conserving surgery. J Cancer Res Clin Oncol 2018;144:1253-63. [Crossref] [PubMed]
- Chen T, Chen J, Zhu Y, et al. CD163, a novel therapeutic target, regulates the proliferation and stemness of glioma cells via casein kinase 2. Oncogene 2019;38:1183-99. [Crossref] [PubMed]
- Vidyarthi A, Agnihotri T, Khan N, et al. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother 2019;68:1995-2004. [Crossref] [PubMed]
- Leblond MM, Pérès EA, Helaine C, et al. M2 macrophages are more resistant than M1 macrophages following radiation therapy in the context of glioblastoma. Oncotarget 2017;8:72597-612. [Crossref] [PubMed]
- Liu YD, Yu L, Ying L, et al. Toll-like receptor 2 regulates metabolic reprogramming in gastric cancer via superoxide dismutase 2. Int J Cancer 2019;144:3056-69. [Crossref] [PubMed]
- Zhao S, Zhang Y, Zhang Q, et al. Toll-like receptors and prostate cancer. Front Immunol 2014;5:352. [Crossref] [PubMed]
- Semlali A, Parine NR, Al-Numair NS, et al. Potential role of Toll-like receptor 2 expression and polymorphisms in colon cancer susceptibility in the Saudi Arabian population. Onco Targets Ther 2018;11:8127-41. [Crossref] [PubMed]
- Qian J, Luo F, Yang J, et al. TLR2 promotes glioma immune evasion by downregulating MHC class II molecules in microglia. Cancer Immunol Res 2018;6:1220-33. [Crossref] [PubMed]
- Li C, Ma L, Liu Y, et al. TLR2 promotes development and progression of human glioma via enhancing autophagy. Gene 2019;700:52-9. [Crossref] [PubMed]
- Gao F, Zhang C, Zhou C, et al. A critical role of toll-like receptor 2 (TLR2) and its’ in vivo ligands in radio-resistance. Sci Rep 2015;5:13004. [Crossref] [PubMed]
- Chang CY, Jeon SB, Yoon HJ, et al. Glial TLR2-driven innate immune responses and CD8+ T cell activation against brain tumor. Glia 2019;67:1179-95. [Crossref] [PubMed]
- Sayegh ET, Bloch O, Parsa AT. Complement anaphylatoxins as immune regulators in cancer. Cancer Med 2014;3:747-58. [Crossref] [PubMed]
- Zhang J, Chen M, Zhao Y, et al. Complement and coagulation cascades pathway correlates with chemosensitivity and overall survival in patients with soft tissue sarcoma. Eur J Pharmacol 2020;879:173121. [Crossref] [PubMed]
- Pan YB, Wang S, Yang B, et al. Transcriptome analyses reveal molecular mechanisms underlying phenotypic differences among transcriptional subtypes of glioblastoma. J Cell Mol Med 2020;24:3901-16. [Crossref] [PubMed]
- Surace L, Lysenko V, Fontana AO, et al. Complement Is a central mediator of radiotherapy-induced tumor-specific immunity and clinical response. Immunity 2015;42:767-77. [Crossref] [PubMed]


