
Identification of adriamycin resistance genes in breast cancer based on microarray data analysis
Introduction
Breast cancer is a common malignant tumor in clinical practice, which occurs in the breast epithelium. Breast cancer is one of the leading life-threatening diseases in women, with the highest incidence rate of 1 million patients and a mortality rate of about 500,000 people (1). In China, breast cancer can cause 279,000 new cases and 66,000 deaths annually (2). Currently, surgery, chemotherapy, and radiotherapy are the therapeutic methods mostly adopted in clinical practice. Early neoadjuvant chemotherapy can significantly improve the progression and prognosis (3). However, despite the advantages, chemotherapy has its limits due to the drug resistance emerged in certain patients (4), and adriamycin (ADR) resistance is the most common one. Therefore, it is of great significance to explore the mechanisms and molecular pathways of ADR resistance in breast cancer.
Nowadays, bioinformatic methods have been widely used in various fields of life sciences, especially in the field of oncology. By using functional genomics and proteomics, researchers can explore the pathogenesis of cancer, as well as the development of screening and targeted drugs, to provide new ideas and theoretical basis for cancer therapy (5). This study adopted bioinformatic techniques, aiming at analyzing gene expression profiles of ADR-resistant breast cancer with public data sources, and screening differentially expressed genes (DEGs) in ADR-resistant and ADR-sensitive cases, and also constructing DEGs-encoded protein-protein interaction (PPI) networks, to analyze and discover the potential genes associated with ADR resistance, and to provide new clues for further researches of the molecular mechanisms and the development of clinical treatment methods.
Methods
Data collection
The gene expression profile dataset was downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (6) (http://www.ncbi.nlm.nih.gov/geo). The GSE76540 dataset (7) was based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, consisting of 3 chemo-sensitive samples and 3 chemo-resistant samples.
Screening for DEGs
The GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/, accessed January 25th, 2019) online tool was employed to detect the DEGs in chemo-sensitive cases and chemo-resistant cases (8), respectively. Adjusted P<0.01, P<0.01 and fold change (FC) ≥2 were considered as the cut-off criterion.
Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs
GO analysis is a tool widely used for biological function annotation of specific genes and gene products, which can be divided into biological process (BP), molecular function (MF), and cellular component (CC) analysis (9). KEGG is a high-throughput database that uses molecular experimental techniques to explain the advanced biological functions of cells and other organisms at the genomic level (10). The GO analysis and KEGG enrichment analysis of DEGs were conducted with the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (https://david.ncifcrf.gov/), and P<0.01 and gene counts >10 were considered statistically significant (11,12).
PPI network construction
The relevant nodes and network diagrams of protein interaction were predicted and analyzed by using the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) (13). We predicted the protein information by uploading the selected differential genes to the STRING database. The protein interaction pairs with combine score greater than 0.4 were extracted and imported into the Cytoscape software (www.cytoscape.org/) (14) to achieve a clear visualization of protein interaction network. At the same time, the degree model of plug-in Cytohubba in Cytoscape software was adopted to evaluate the importance of each protein node and the overall contribution to the protein network. The 15 top-rated genes selected by degree method were regarded as hub genes.
Survival analysis of hub genes
GEPIA (http://gepia.cancer-pku.cn/detail.php) (15) is a public database based on tumor analysis, providing freely published tumor gene transcriptome data [including the Cancer Genome Atlas (TCGA) database], and also collecting and summarizing a large number of tumor-related gene expression levels and patient survival information. Herein, the impact of key tumor genes on the survival and prognosis were described based on the GEPIA database.
Results
Identification of DEGs
A total of 1,481 DEGs were achieved after analyzing the GSE76540 dataset by using the GEO2R online tool, including 549 up-regulated genes and 932 down-regulated genes. The five genes with the most significant up-regulation were MMP1, TMEM200A, NEFH, KRTAP2-4, and PPP1R14A, while the most significant down-regulated genes were SYTL5, MAL2, WISP2, GREB1, and FXYD3 (Table 1).
Table 1
| Expression | Genes | Adjust P value | LogFC |
|---|---|---|---|
| Up-regulation | MMP1 | 1.02E−4 | 9.9795696 |
| TMEM200A | 1.02E−4 | 9.2300751 | |
| NEFH | 1.02E−4 | 9.6770347 | |
| KRTAP2-4 | 1.02E−4 | 11.1822447 | |
| PPP1R14A | 1.02E−4 | 8.9519095 | |
| Down-regulation | SYTL5 | 1.02E−4 | −10.4814297 |
| MAL2 | 1.02E−4 | −9.3188679 | |
| WISP2 | 1.2E−4 | −8.6980133 | |
| GREB1 | 1.48E−4 | −6.064777 | |
| FXYD3 | 1.48E−4 | −8.8321929 |
FC, fold change.
Functional enrichment analysis of DEGs
The results of GO analysis indicated that the five BPs with the most up-regulated genes were: extracellular matrix organization, positive regulation of transcription from RNA polymerase II promoter, lung development, positive regulation of gene expression, axon guidance, while the BPs with the most down-regulated genes included: homophilic cell adhesion via plasma membrane adhesion molecules, positive regulation of transcription from RNA polymerase II promoter, signal transduction, cell-cell signaling, and angiogenesis (Table 2). The CC analysis showed that the up-regulated genes were mostly enriched in plasma membrane, extracellular space, extracellular region, basement membrane, and cell surface, in contrast of which, the down-regulated genes were mainly in extracellular exosome, bicellular tight junction, plasma membrane, integral component plasma membrane, and extracellular space (Table 3). According to the MF analysis, the most up-regulated genes could be detected in heparin binding, extracellular matrix structural constituent, signal transducer activity, transcription factor activity, sequence-specific DNA binding, and RNA polymerase II core promoter proximal region sequence-specific binding, whereas the down-regulated genes were enriched significantly in calcium ion binding, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, and sequence-specific binding (Table 4).
Table 2
| Expression | Term | Count | P value | Benjamin |
|---|---|---|---|---|
| Up-regulation | Extracellular matrix organization | 24 | 6.9E−8 | 1.8E−4 |
| Positive regulation of transcription from RNA polymerase II promoter | 57 | 1.4E−5 | 1.8E−2 | |
| Lung development | 12 | 2.6E−5 | 2.2E−2 | |
| Positive regulation of gene expression | 22 | 9.9E−5 | 6.2E−2 | |
| Axon guidance | 15 | 5.5E−4 | 2.1E−1 | |
| Down-regulation | Homophilic cell adhesion via plasma membrane adhesion molecules | 15 | 2.9E−6 | 5.8E−3 |
| Positive regulation of transcription from RNA polymerase II promoter | 36 | 4.8E−4 | 1.5E−1 | |
| Signal transduction | 38 | 2.5E−3 | 4.3E−1 | |
| Cell-cell signaling | 13 | 4.6E−3 | 4.8E−1 | |
| Angiogenesis | 12 | 4.7E−3 | 4.7E−1 |
Table 3
| Expression | Term | Count | P value | Benjamin |
|---|---|---|---|---|
| Up-regulation | Plasma membrane | 179 | 3.2E−7 | 1.2E−4 |
| Extracellular space | 75 | 8.1E−7 | 1.5E−4 | |
| Extracellular region | 84 | 2.1E−6 | 2.5E−4 | |
| Basement membrane | 13 | 5.1E−6 | 4.6E−4 | |
| Cell surface | 36 | 3.3E−5 | 2.4E−3 | |
| Down-regulation | Extracellular exosome | 83 | 5.4E−5 | 1.4E−2 |
| Bicellular tight junction | 11 | 6.6E−5 | 8.7E−3 | |
| Plasma membrane | 109 | 2.3E−4 | 1.5E−2 | |
| Integral component plasma membrane | 44 | 2.0E−4 | 9.8E−3 | |
| Extracellular space | 41 | 4.2E−3 | 1.5E−1 |
Table 4
| Expression | Term | Count | P value | Benjamin |
|---|---|---|---|---|
| Up-regulation | Heparin binding | 16 | 1.3E−4 | 8.6E−2 |
| Extracellular matrix structural constituent | 11 | 2.1E−4 | 6.7E−2 | |
| Signal transducer activity | 18 | 2.1E−4 | 4.5E−2 | |
| Transcription factor activity sequence-specific DNA binding | 51 | 2.2E−4 | 3.6E−2 | |
| RNA polymerase II core promoter proximal region sequence-specific binding | 18 | 1.1E−3 | 1.2E−1 | |
| Down-regulation | Calcium ion binding | 38 | 6.7E−8 | 3.4E−5 |
| RNA polymerase II core promoter proximal region sequence-specific binding | 20 | 2.0E−7 | 4.9E−5 | |
| RNA polymerase II core promoter proximal region sequence-specific binding | 16 | 4.3E−3 | 4.1E−1 | |
| Sequence-specific binding | 20 | 6.5E−3 | 4.8E−1 |
In addition, the KEGG pathway enrichment analysis suggested that the DEGs were enriched in: cancer pathways, PI3K AKT signaling pathway, focal adhesion, Ras signaling pathway, cytokine-cytokine receptor interaction, MAPA signaling pathway, hematopoietic cell lineage, amoebiasis, calcium signaling pathway, oxytocin signaling pathway, and proteoglycans in cancer (Table 5).
Table 5
| Term | Count | P value | Benjamin |
|---|---|---|---|
| Pathways in cancer | 31 | 1.7E−6 | 3.8E−4 |
| Focal adhesion | 19 | 4.0E−5 | 4.6E−3 |
| Hematopoietic cell lineage | 11 | 2.5E−4 | 1.9E−2 |
| Amoebiasis | 11 | 1.2E−3 | 6.8E−2 |
| Calcium signaling pathway | 14 | 2.7E−3 | 1.2E−1 |
| Ras signaling pathway | 16 | 3.1E−3 | 1.1E−1 |
| PI3K AKT signaling pathway | 21 | 3.3E−3 | 1.0E−1 |
| Oxytocin signaling pathway | 12 | 5.2E−3 | 1.4E−1 |
| Cytokine-cytokine receptor interaction | 16 | 6.1E−3 | 1.3E−1 |
| Proteoglycans in cancer | 14 | 6.8E−3 | 1.3E−1 |
| MAPA signaling pathway | 16 | 8.7E−3 | 1.4E−1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
PPI network and hub genes identification
A total of 635 pairs of interacting proteins were achieved after screening and the network structure was constructed (Figure 1). Fifteen key genes were obtained by using the degree model of plug-in Cytohubba in Cytoscape software, which were considered as hub genes, including: CDH1, ESR1, SOX2, AR, GATA3, FOXA1, KRT19, CLDN7, AGR2, ESRP1, RAB25, CLDN4, IGF1R, CLDN3, and IRS1 (Table 6, Figure 2).

Table 6
| Gene | Degree | MCC | MNC |
|---|---|---|---|
| CDH1 | 50 | 1,817 | 43 |
| ESR1 | 41 | 1,142 | 37 |
| AR | 24 | 430 | 20 |
| SOX2 | 24 | 309 | 21 |
| FOXA1 | 23 | 1,028 | 23 |
| GATA3 | 23 | 762 | 23 |
| AGR2 | 18 | 229 | 17 |
| CLDN7 | 18 | 1,314 | 18 |
| KRT19 | 18 | 372 | 15 |
| ESRP1 | 16 | 922 | 14 |
| IGF1R | 15 | 114 | 13 |
| CLDN4 | 15 | 1,256 | 13 |
| RAB25 | 15 | 766 | 11 |
| IRS1 | 14 | 98 | 12 |
| CLDN3 | 14 | 1,284 | 14 |
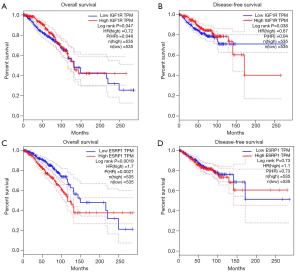
Survival analysis of hub genes
After analyzing influences of the 15 hub genes on the overall survival (OS) and disease-free survival (DFS) prognosis, the results suggested that only the insulin like growth factor 1 receptor (IGF1R) gene had significant impact on both OS and DFS (log-rank P=0.047, 0.038 respectively), and the epithelial splicing regulatory protein 1 (ESRP1) gene showed significant effect on the OS only (log-rank P=0.0019). The rest of hub genes did not have significant influence on the survival prognosis (Figure 3).
Discussion
ADR is a first-line neoadjuvant chemotherapy drug for breast cancer (16,17). Although it is effective, long-term use of ADR can always cause drug resistance. The mechanisms of drug resistance in tumor cells have not been completely clarified. However, theories have been established that tumor chemotherapy resistance is caused by a combination of multi-drug resistance pump and enzymes. In addition, genetic differences between individuals may also contribute to the drug resistance to some extent. In order to improve the efficacy of chemotherapy for breast cancer, it is necessary to explore the potential mechanisms of drug resistance.
It is widely believed that glycoprotein P-gp and multi-drug resistance protein MRP1 are involved in the resistance to several drugs in tumor cells. The catalytic ATP pumps can generate energy to expel the drugs out of cells, thus reducing the effective intracellular concentration, and reducing the inhibition effect on tumor cells, which is manifested as drug resistance in clinical practice (18-23). In addition, a series of genes associated with ADR resistance were detected, such as Nrf2, SOX2, SPIN1, COP1, Mdr1 and so on (24-29). In this study, we explored ADR resistance genes in breast cancer with the GSE76540 dataset. By analyzing 3 drug-resistant samples and 3 drug-sensitive samples, overall 1,481 DEGs were detected, including 932 down-regulated genes and 549 up-regulated genes. GO functional analysis and KEGG pathway analysis were conducted to obtain biological characteristics of the selected genes for further comprehensive analysis.
The GO analysis suggested that the BPs with the most enriched DEGs included: extracellular matrix organization, positive regulation of transcription from RNA polymerase II promoter, lung development, positive regulation of gene expression, axon guidance, homophilic cell adhesion via plasma membrane adhesion molecules, positive regulation of transcription from RNA polymerase II promoter, signal transduction, cell-cell signaling, and angiogenesis. The MF analysis showed that enriched DEGs can be detected in certain MFs, such as heparin binding, extracellular matrix structural constituent, signal transducer activity, transcription factor activity, sequence-specific DNA binding, RNA polymerase II core promoter proximal region sequence-specific binding, calcium ion binding, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, RNA polymerase II core promoter proximal region sequence-specific binding, sequence-specific binding and so on. The CC analysis indicated that the DEGs were mostly enriched in plasma membrane, extracellular space, extracellular region, basement membrane, cell surface, extracellular exosome, bicellular tight junction, plasma membrane, integral component plasma membrane, and extracellular space.
The KEGG pathway analysis showed that DEGs were mainly enriched in cancer-associated pathways such as pathways in cancer, PI3K AKT signaling pathway, focal adhesion, Ras signaling pathway, cytokine-cytokine receptor interaction, MAPA signaling pathway, hematopoietic cell lineage, amoebiasis, calcium signaling pathway, oxytocin signaling pathway, and proteoglycans in cancer.
According to the PPI network and the analysis with the degree model, 15 genes with a high degree of association were selected as hub genes, among which CDH1, ESR1, SOX2, AR, and GATA3 were the top five genes. The CDH1 gene is a member of the cadherin superfamily, which has been demonstrated to be correlated with a series cancer occurring in different parts, such as gastric cancer, breast cancer, colorectal cancer, thyroid cancer, ovarian cancer and so on. Mutation of the CDH1 gene can promote the progression and growth of tumor tissues, as a result of which, it may be related to drug resistance in tumor cells (30). The ESR1 gene encodes an estrogen receptor involved in the pathological process of breast cancer. The SOX2 gene plays an important role in the regulation of stem cell growth and development. The AR gene encodes an androgen receptor that promotes androgen binding, and its mutation can lead to loss of control of tumor cells (31). The GATA3 gene is a transcription factor-regulated protein whose mutation can lead to developmental disorders of immune cells thus damaging to the immune system (32).
In the prognostic survival analysis, only two genes were related with prognostic significance. The IGF1R gene had influence on both OS and DFS, while the ESRP1 gene only affected the OS. Therefore, patients screened for these two genes may have worse prognosis and quality of life. It is necessary to modify the chemotherapeutic drugs in advance and redesign the chemotherapeutic regimen.
Conclusions
In summary, this study contributed to a better understanding of the molecular screening and potential mechanisms of drug resistance in breast cancer. By using bioinformatic techniques, 15 hub genes were selected among a total of 1,481 DEGs. The IGF1R and ESRP1 genes might be options as prognostic biomarkers for breast cancer. Further studies are needed to verify the results.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2145). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wei C, Jiang L. Application value of mammography and color Doppler ultrasonography combined with MRI for early diagnosis of breast cancer. China Modern Medicine 2017;24:70-2.
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett 2017;401:63-71. [Crossref] [PubMed]
- Kaufmann M, von Minckwitz G, Mamounas EP, et al. Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol 2012;19:1508-16. [Crossref] [PubMed]
- DeMichele A, Yee D, Esserman L. Mechanisms of Resistance to Neoadjuvant Chemotherapy in Breast Cancer. N Engl J Med 2017;377:2287-9. [Crossref] [PubMed]
- Kato M. Bioinformatics in Cancer Clinical Sequencing -- An Emerging Field of Cancer Personalized Medicine. Gan To Kagaku Ryoho 2016;43:391-7. [PubMed]
- Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol 2016;1418:93-110. [Crossref] [PubMed]
- Wang C, Jin H, Wang N, et al. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3beta/beta-catenin Signaling. Theranostics 2016;6:1205-19. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- D'Hondt V, Canon JL, Roca L, et al. UCBG 2-04: Long-term results of the PACS 04 trial evaluating adjuvant epirubicin plus docetaxel in node-positive breast cancer and trastuzumab in the human epidermal growth factor receptor 2-positive subgroup. Eur J Cancer 2019;122:91-100. [Crossref] [PubMed]
- Zhang P, He D, Chen Z, et al. Chemotherapy enhances tumor vascularization via Notch signaling-mediated formation of tumor-derived endothelium in breast cancer. Biochem Pharmacol 2016;118:18-30. [Crossref] [PubMed]
- Monk BC, Keniya MV, Sabherwal M, et al. Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161. Antimicrob Agents Chemother 2018;63:e02114-18. [Crossref] [PubMed]
- Sarathy JP, Gruber G, Dick T. Re-Understanding the Mechanisms of Action of the Anti-Mycobacterial Drug Bedaquiline. Antibiotics (Basel) 2019;8:E261. [Crossref] [PubMed]
- Lee CA, O'Connor MA, Ritchie TK, et al. Breast cancer resistance protein (ABCG2) in clinical pharmacokinetics and drug interactions: practical recommendations for clinical victim and perpetrator drug-drug interaction study design. Drug Metab Dispos 2015;43:490-509. [Crossref] [PubMed]
- Su W, Kumar V, Ding Y, et al. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc Natl Acad Sci U S A 2018;115:5157-62. [Crossref] [PubMed]
- Wang L, Liu L, Chen Y, et al. Correlation between adenosine triphosphate (ATP)-binding cassette transporter G2 (ABCG2) and drug resistance of esophageal cancer and reversal of drug resistance by artesunate. Pathol Res Pract 2018;214:1467-73. [Crossref] [PubMed]
- Pan X, Yang X, Zang J, et al. Downregulation of eIF4G by microRNA-503 enhances drug sensitivity of MCF-7/ADR cells through suppressing the expression of ABC transport proteins. Oncol Lett 2017;13:4785-93. [Crossref] [PubMed]
- Na HK, Surh YJ. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free Radic Biol Med 2014;67:353-65. [Crossref] [PubMed]
- Zeng H, Wang L, Wang J, et al. microRNA-129-5p suppresses Adriamycin resistance in breast cancer by targeting SOX2. Arch Biochem Biophys 2018;651:52-60. [Crossref] [PubMed]
- Chen X, Wang YW, Gao P. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res 2018;37:100. [Crossref] [PubMed]
- Wan L, Tian Y, Zhang R, et al. MicroRNA-103 confers the resistance to long-treatment of adriamycin to human leukemia cells by regulation of COP1. J Cell Biochem 2018;119:3843-52. [Crossref] [PubMed]
- Jagadeeshan S, David D, Jisha S, et al. Solanum nigrum Unripe fruit fraction attenuates Adriamycin resistance by down-regulating multi-drug resistance protein (Mdr)-1 through Jak-STAT pathway. BMC Complement Altern Med 2017;17:370. [Crossref] [PubMed]
- Wei Y, Liu D, Jin X, et al. PA-MSHA inhibits the growth of doxorubicin-resistant MCF-7/ADR human breast cancer cells by downregulating Nrf2/p62. Cancer Med 2016;5:3520-31. [Crossref] [PubMed]
- Corso G, Intra M, Trentin C, et al. CDH1 germline mutations and hereditary lobular breast cancer. Fam Cancer 2016;15:215-9. [Crossref] [PubMed]
- Ricciardi GR, Adamo B, Ieni A, et al. Androgen Receptor (AR), E-Cadherin, and Ki-67 as Emerging Targets and Novel Prognostic Markers in Triple-Negative Breast Cancer (TNBC) Patients. PLoS One 2015;10:e0128368. [Crossref] [PubMed]
- Wan YY. GATA3: a master of many trades in immune regulation. Trends Immunol 2014;35:233-42. [Crossref] [PubMed]


