
Druggable driver gene alterations in redefined large cell carcinoma in Chinese patients: an observational study
Introduction
Large cell carcinoma (LCC) is the third most common subtype of non-small-cell lung cancer (NSCLC) after lung adenocarcinoma (LUAD) and squamous cell carcinoma (SCC). According to the 2015 World Health Organization (WHO) classification of lung tumors, the diagnosis of LCC has been restricted only to “resected tumors that lack any clear morphologic or immunohistochemical (IHC) differentiation towards LUAD, SCC, or small cell carcinoma” (1).
The updated classification highlights the significance of IHC staining in tumor classification. After IHC analysis, the resected and undifferentiated NSCLCs with immunopositivity for LUAD markers [thyroid transcription factor-1 (TTF-1) or napsin A], or squamous markers (p40, p63, or CK5/6), are now reclassified as solid LUADs or non-keratinizing SCC. Uncommon specific cancer types, such as large cell neuroendocrine carcinoma, basaloid carcinoma, and lymphoepithelioma-like carcinoma, are no longer included in the LCC category. Only those with marker-null phenotype or unclear immunophenotypes are currently classified as LCC (1,2).
Although the updated classification of lung cancer has led to an enormous change in the histological type of LCC, the treatment strategy for LCC remains unchanged. Compared with molecular changes in patients with LUAD, who benefit largely from treatments targeting epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) translocations, the genomic alterations in redefined LCC have yet to be completely characterized (3,4). Due to the lack of an effective targeted therapy for LCC, chemotherapy remains the first-line treatment for patients with LCC (5,6).
Few reports have investigated the genetic status of LCC in Chinese patients under the 2015 WHO classification. Therefore, in this study we aimed to analyze the distribution of druggable driver gene alterations, including EGFR, Kirsten rat sarcoma 2 viral oncogene homolog (KRAS), proto-oncogene B-Raf (BRAF), and phosphatidylinositol-4,5 biphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutations, and echinoderm microtubule-associated protein-like 4 (EML4)-ALK, and ROS proto-oncogene 1 (ROS1) translocations, and their clinical features in a large population of patients with LCC who underwent therapeutic resection. Additionally, we assessed the clinical outcomes of LCC patients with or without gene alterations. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1675).
Methods
Study design
We conducted a retrospective cohort study by manually reviewing and extracting data from the electronic medical records of patients diagnosed with LCC at the Shanghai Pulmonary Hospital, Affiliated to the Tongji University in China, between June 2015 and December 2018. Patients’ clinical characteristics, driver gene mutation status, and disease-free survival (DFS) were determined.
Patient cohort
A consecutive cohort of 322 patients was included in this study. All patients underwent surgical resection at the Shanghai Pulmonary Hospital, Affiliated to the Tongji University in China, between June 2015 and December 2018. All tumors were diagnosed according to the 2015 WHO criteria and staged according to the seventh edition of the tumor-node-metastasis (TNM) system. The study was conducted in accordance with the tenets of the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board at the Shanghai Pulmonary Hospital (FK19-177), and informed consent was provided by all participants.
Driver gene mutation analysis
The amplification-refractory mutation system (ARMS) was used as the molecular diagnostic method in our study. Genomic DNA and total RNA were extracted from five serial slices of a 5-µm paraffin section using the DNA FFPE Tissue Kit and RNeasy Kit (Qiagen, Hilden, Germany). Mutations in EGFR, KRAS, BRAF, and PIK3CA genes, as well as EML4-ALK and ROS1 translocations were detected using ACCB Diagnostics Kit (ACCB Biotech Ltd., Beijing, China), according to the manufacturer’s protocol (7). The test could detect mutations at a sensitivity of 1% in no less than 5 ng/µL of DNA and RNA samples.
Groupings
Patients with redefined LCC were divided into two groups. The mutation group included all cases with driver gene mutations identified in the current study, including EGFR, KRAS, and PIK3CA mutations, and EML4-ALK translocations. Patients with mutations other than the ones mentioned above were assigned to the wild-type (WT) group.
Clinical data collection
Data regarding participant characteristics at the time of lung cancer diagnosis, including age, sex, smoking history, tumor size and site, pathological TNM stage, and histological type were collected retrospectively. Calculation of DFS was from the date of surgical resection until the date of confirmed recurrence from any cause.
Follow-up
At the time of surgical hospitalization, all patients were asked to provide their personal telephone number and that of a contact person to be used during active follow-up. Patients with redefined LCC were contacted if they had not visited the clinic for 3 months after the date of the last visit. Patients were considered lost to follow-up if they had not visited the clinic for 6 months, and telephone contact was not achieved at least twice on 2 separate days.
Statistical analysis
Statistically significant differences in categorical variables between the groups were analyzed using the chi-squared (χ2) or Fisher’s exact test, as appropriate. The Kaplan-Meier method was used to estimate DFS, and the log-rank test for univariate analysis. The Cox proportional hazards model was used for multivariate analysis. The covariates considered for multivariate analysis were gender, age, TNM stage, tumor size, and smoking status. No missing data were observed in the current study. All tests were two-sided, and a P value <0.05 was defined as statistically significant. Statistical analyses were carried out using SPSS version 20.0 (SPSS Statistics, IBM, Chicago, IL, USA).
Results
Redefinition of LCC by hematoxylin and eosin (HE) staining and IHC analysis
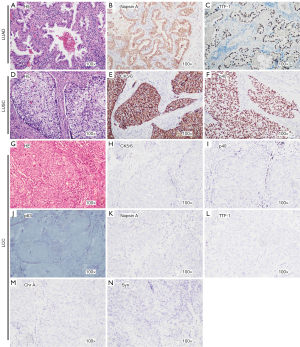
According to the 2015 WHO criteria, negative IHC staining for TTF-1, napsin A, chromogranin A (Chr A), synaptophysin (Syn), p40, p63, and CK5/6 in a resected specimen was defined as LCC. Representative samples with immunopositivity for each marker and typical HE staining of LCCs are shown in Figure 1. We included IHC staining of LUAD and lung squamous cell carcinoma (LUSC) tissue sections to serve as positive controls (Figure 1). Moreover, HE and IHC staining for 1 case of KRAS-mutated LCC was performed (Figure S1) to demonstrate that driver gene alteration positive samples were not LUAD or LUSC specimens.
An overview of clinical features in LCC patients
As presented in Figure 2, of 322 patients, 192 were redefined as LCC and included in the analysis. The clinicopathological data are summarized in Table 1. The proportion of males was much higher than that of females (94.3% vs. 5.7%). Detection of LCC was more frequent in patients >60 years of age (71.4%). Of the 192 cases, 119 (62%) were stage I, 35 (18.2%) were stage II, and 38 (19.8%) were stage III. There was no significant difference in smoking status or tumor size and site.
Table 1
| Characteristics | N (%) |
|---|---|
| Sex | |
| Female | 11 (5.7) |
| Male | 181 (94.3) |
| Age, years | |
| Mean [range] | 64.74 [39–80] |
| Median | 65.5 |
| ≤60 (median =56) | 55 (28.6) |
| >60 (median =69) | 137 (71.4) |
| Smoking | |
| Non-smoker | 98 (51.0) |
| Smoker | 94 (49.0) |
| TNM stage (male/female) | |
| I (113/6) | 119 (62.0) |
| II (33/2) | 35 (18.2) |
| III A (35/3) | 38 (19.8) |
| TNM stage (non-smoker/smoker) | |
| I (57/62) | 119 (62.0) |
| II (20/15) | 35 (18.2) |
| III A (21/17) | 38 (19.8) |
| Tumor size | |
| ≤3 cm | 92 (47.9) |
| >3 cm | 100 (52.1) |
| Site | |
| Left | 98 (51.0) |
| Right | 94 (49.0) |
The clinical features of 192 patients with LCC are summarized. LCC, large cell carcinoma; TNM, tumor-node-metastasis.
Driver gene mutation profile in patients with LCC
Mutations were identified in 27 of the 192 cases. As shown in Table 2, the common driver gene mutation observed in the cohort included 7 cases (3.6%) of EGFR mutations, 1 (0.52%) of EML4-ALK translocation, 15 (7.8%) of KRAS mutations, and 4 (2.1%) of PIK3CA mutations. No alterations were identified in ROS1 and BRAF.
Table 2
| Genes | N (%) |
|---|---|
| EGFR | |
| Wide type | 185 (96.4) |
| Mutation | 7 (3.6) |
| EML4-ALK | |
| Wide type | 191 (99.5) |
| Mutation | 1 (0.5) |
| KRAS | |
| Wide type | 177 (92.2) |
| Mutation | 15 (7.8) |
| ROS1 | |
| Wide type | 192 (100.0) |
| Mutation | 0 |
| BRAF | |
| Wide type | 192 (100.0) |
| Mutation | 0 |
| PIK3CA | |
| Wide type | 188 (97.9) |
| Mutation | 4 (2.1) |
LCC, large cell carcinoma; EGFR, epidermal growth factor receptor; EML4-ALK, translocations in echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase; KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; ROS1, ROS proto-oncogene 1; BRAF, proto-oncogene B-Raf; PIK3CA, phosphatidylinositol-4,5 biphosphate 3-kinase catalytic subunit alpha.
Clinical features of mutated LCC
The distribution and clinical features of the 27 cases harboring driver gene mutations are listed in Table 3. Of the 7 cases harboring EGFR mutations, including 6 cases of EGFR L858R, and 1 of EGFR 19-del, 5 were male, and 2 of them had a history of smoking. All the cases in the EML4-ALK translocation, KRAS, and PIK3CA mutations group were male. Of the 15 cases in the KRAS mutation group, 9 were smokers. The mutation type observed in the cohort of 15 patients with KRAS mutations included KRAS G12C (7/15), KRAS G12D (5/15), and KRAS G12V (3/15). Of the 4 cases in the PIK3CA mutation group, only 1 was a smoker. Only 1 case, a current smoker, was identified in the EML4-ALK translocation group.
Table 3
| No. | Mutation type | Sex | Age | Smoking | TNM stage | Tumor size | Site |
|---|---|---|---|---|---|---|---|
| 1 | EGFR-L858R | Male | 64 | Non-smoker | I | <3 cm | Right |
| 2 | EGFR-L858R | Male | 72 | Non-smoker | III | >3 cm | Left |
| 3 | EGFR-L858R | Female | 70 | Non-smoker | I | >3 cm | Left |
| 4 | EGFR-L858R | Female | 80 | Non-smoker | II | >3 cm | Right |
| 5 | EGFR-L858R | Male | 54 | Current smoker | I | <3 cm | Right |
| 6 | EGFR-L858R | Male | 74 | Current smoker | I | <3 cm | Left |
| 7 | EGFR-19del | Male | 45 | Non-smoker | II | >3 cm | Left |
| 8 | KRAS-G12C | Male | 70 | Non-smoker | I | <3 cm | Right |
| 9 | KRAS-G12C | Male | 63 | Current smoker | I | <3 cm | Left |
| 10 | KRAS-G12C | Male | 72 | Non-smoker | I | <3 cm | Left |
| 11 | KRAS-G12D | Male | 65 | Non-smoker | I | <3 cm | Right |
| 12 | KRAS-G12C | Male | 54 | Non-smoker | II | <3 cm | Right |
| 13 | KRAS-G12V | Male | 52 | Current smoker | I | >3 cm | Right |
| 14 | KRAS-G12D | Male | 57 | Non-smoker | I | <3 cm | Right |
| 15 | KRAS-G12D | Male | 50 | Previous smoker | I | <3 cm | Right |
| 16 | KRAS-G12C | Male | 67 | Non-smoker | II | <3 cm | Right |
| 17 | KRAS-G12V | Male | 75 | Current smoker | I | >3 cm | Right |
| 18 | KRAS-G12D | Male | 69 | Current smoker | II | <3 cm | Left |
| 19 | KRAS-G12C | Male | 72 | Current smoker | I | <3 cm | Left |
| 20 | KRAS-G12V | Male | 63 | Current smoker | I | <3 cm | Right |
| 21 | KRAS-G12C | Male | 71 | Current smoker | I | >3 cm | Left |
| 22 | KRAS-G12D | Male | 62 | Current smoker | I | <3 cm | Right |
| 23 | ALK-EA2 | Male | 66 | Current smoker | I | >3 cm | Left |
| 24 | PI3K-E545K | Male | 69 | Non-smoker | I | <3 cm | Right |
| 25 | PI3K-E454K | Male | 56 | Non-smoker | II | >3 cm | Left |
| 26 | PI3K-E545K | Male | 46 | Non-smoker | II | >3 cm | Left |
| 27 | PI3K-E542K | Male | 74 | Current smoker | I | <3 cm | Left |
The distribution and clinical features of the 27 cases harboring driver gene mutations are listed in this table. LCC, large cell carcinoma; EGFR L858R, an amino acid substitution of the leucine at position 858 by an arginine at exon 21 in EGFR; EGFR-19del, EGFR exon 19 deletion; KRAS-G12C, a single point mutation with a glycine-to-cysteine substitution at exon 12 in KRAS; KRAS-G12V, a single point mutation with a glycine-to-valine substitution at exon 12 in KRAS; KRAS-G12D, a single point mutation with a glycine-to-aspartic acid substitution at exon 12 in KRAS; ALK-EA2, EML4-ALK translocations exhibiting fusions between exon 20 of EML4 and exon 20 of ALK; PI3K-E454K, a single amino-acid substitution at glutamine 454 to lysine in the activating segment of PI3K; PI3K-E545K, a single amino-acid substitution at glutamine 545 to lysine in the activating segment of PI3K; PI3K-E542K, a single amino-acid substitution at glutamine 542 to lysine in the activating segment of PI3K.
Analysis of DFS
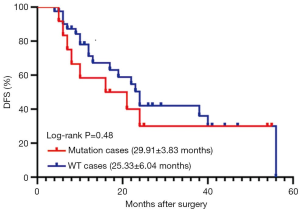
We divided all the participants with redefined LCC into two groups. The mutation group included cases with all driver gene mutations identified in the current study, including EGFR, KRAS, and PIK3CA mutations, and EML4-ALK translocation. Patients with mutations other than those mentioned above were assigned to the WT group. As shown in Figure 3, no significant difference in DFS was identified between these two groups (29.911±3.826 vs. 25.333±6.035 months, P=0.476). Since KRAS mutation was the most common mutation in LCC, we then examined whether the presence of KRAS mutation alone could lead to reduced DFS. Our analysis demonstrated that there was no significant difference in DFS between the KRAS-positive patients and non KRAS-positive patients (P=0.232).
Discussion
In the present study, the druggable driver gene mutation profiles and clinical outcomes were evaluated in a large cohort of postoperative patients with LCC. Detection of LCC was more frequent in elderly male patients, and the prognosis for all LCC patients was poor. The overall mutation rates of driver genes in LCC, including mutations in EGFR, KRAS, EML4-ALK, and PIK3CA, were lower in LCC than in LUAD.
Evaluation of the clinical characteristics of the LCC participants in this study revealed no differences in smoking status or tumor size and site. However, LCC was more frequently detected in males and in patients aged ≥60 years, consistent with the results of a study from another center in China (8). Mutation in EGFR occurred more frequently in both male and female non-smokers, whereas KRAS mutation occurred solely in males, and more frequently in smokers. EML4-ALK translocation and PIK3CA mutation were also observed in males, and patients with PIK3CA mutation tended to be smokers.
In our study, the incidence of EGFR mutation in patients with LCC was 3.6%, which was much lower than that reported in patients with LUAD (9-11). An L858R mutation in exon 21 of the EGFR gene is reportedly the most common type of mutation in patients with LUAD (9,10). Consistently, EGFR L858R mutations accounted for 85.7% of common EGFR mutations in the current report, and remained the dominant EGFR mutation in LCC. However, the uncommon EGFR mutations reported in LUAD, including 20-ins and T790M, were not observed in LCC (12,13).
Mutations that activate EGFR, including exon 19 deletions or exon 21 L858R, exhibit favorable outcomes after treatment with EGFR-TKI (3,14,15). In our study, 1 of 7 patients with EGFR-tyrosine kinase inhibitor (TKI)-sensitive EGFR mutations received EGFR-TKI (gefitinib) as the first-line treatment (data not shown here). However, the patient showed no response to gefitinib therapy, suggesting that the therapeutic effect of EGFR-TKI might be related to the histological subtype of tumor tissues.
KRAS mutations are the second most frequent oncogenic aberrations in LUAD, with typical mutation rates of around 10% in Asians and up to 30% in Caucasians (11,16). In this study, the rate of KRAS mutations was 7.8%, remaining the most frequent in patients with LCC, which is consistent with previous findings (8,17). Despite its prevalence, mutant KRAS has endured as an intractable therapeutic target, even after decades of extensive effort (18). Recently, pharmacokinetic and pharmacodynamic improvements of direct G12C inhibitors (ARS-1620, AMG 510, and MRTX849) have raised great excitement, and MRTX849 and AMG 510 are currently being tested in first-in-human clinical trials (NCT03785249 and NCT03600883, respectively) (19,20). However, the therapeutic potential of these inhibitors can be impaired by the intrinsic resistance mechanism, suggesting the need for drug combination strategies (21,22). Indeed, CK2 (a catalytic sub-unit encoded by CSNK2A1) was reported as a promising co-target for overcoming MEK/ERK inhibitor resistance in patients with LUAD with KRAS (G12C) mutation (23). The combination of ARS-1620 with PIK3 inhibitors could maximize the response rate and reduce the development of adaptive resistance mechanisms in vitro and in vivo (21). Nonetheless, as the majority of the data were generated from patients with LUAD, the therapeutic efficacy of these inhibitors in patients with LCC warrants further studies.
Mutations in the PIK3CA gene occur less frequently, at about 2–5%, and are prevalent in squamous NSCLC (24,25). In the current study, 4 patients presented the PIK3CA mutation at a rate of 2.1%, similar to that in squamous NSCLC. The PIK3CA mutation has been reported to occur in parallel with other oncogenic driver mutations, and it has also been discovered in EGFR-mutant NSCLCs that have developed acquired resistance to EGFR-TKIs, possibly representing escape mechanisms from the TKI inhibition (26-28). Several small molecules targeting PIK3CA have been evaluated in multiple clinical trials, either in monotherapy or in combination strategy. Data from these clinical trials demonstrated that single-agent PI3K inhibitors offer limited, if any, activity, while combination with other targeted agents and/or cytotoxic chemotherapy may prove to be more efficacious (28).
In our study, EML4-ALK translocation was detected in 1 patient with LCC. Although targeting EML4-ALK translocation with ALK inhibitors has become the cornerstone of managing advanced non-squamous NSCLC harboring EML4-ALK translocation, the relevance of ALK inhibitors in LCC is poorly defined (4).
In terms of prognosis, the median DFS observed in our study was 29.911 and 25.333 months in WT and mutation groups, respectively. These DFS periods were much shorter than those observed in lung SCC at the same center, which were 45.9 and 49.5 months in EGFR-positive and EGFR-negative patients, respectively (29).
This study had several limitations. First, despite the inclusion of large-scale data compared with those of previous studies, our results were produced from patients admitted at a single institution, which can lead to a participant selection bias. Second, due to the relatively low frequency of EGFR mutations in LCC and limited number of patients who accepted the EGFR-TKI therapy, we could not draw definite conclusions concerning the efficacy of EGFR-TKIs in LCC patients harboring EGFR mutations.
Overall, LCC was more frequently detected in elderly male patients with inferior prognoses under the 2015 WHO criteria. The most frequent gene mutations observed in LCC were those of EGFR and KRAS. Activating EGFR mutation occurred more frequently in non-smokers of both genders, whereas KRAS mutation occurred solely in males and more frequently in smokers. The PIK3CA mutations and EML4-ALK translocations were rare in patients with LCC. Our data revealed that the identification of clinically actionable molecular alterations in LCC could help in the research and development of targeted drugs for treating LCC, and may guide personalized cancer treatment decisions in the future.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1675
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1675
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1675
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1675). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the tenets of the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board at the Shanghai Pulmonary Hospital (FK19-177), and informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Micke P, Mattsson JS, Djureinovic D, et al. The Impact of the Fourth Edition of the WHO Classification of Lung Tumours on Histological Classification of Resected Pulmonary NSCCs. J Thorac Oncol 2016;11:862-72.
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med 2006;355:2542-50. [Crossref] [PubMed]
- Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Wang X, Gao Y, Wang B, et al. Analytic and Clinical Validation of an Ultrasensitive, Quantitative Polymerase Chain Reaction Assay for EGFR Mutation Analysis With Circulating Tumor DNA. Arch Pathol Lab Med 2017;141:978-84. [Crossref] [PubMed]
- Liu R, Liu J, Shi T, et al. Clinicopathological and genetic characteristics of pulmonary large cell carcinoma under 2015 WHO classification: a pilot study. Oncotarget 2017;8:100754-63. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Castellanos E, Feld E, Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:612-23. [Crossref] [PubMed]
- Wen YS, Cai L, Zhang XW, et al. Concurrent oncogene mutation profile in Chinese patients with stage Ib lung adenocarcinoma. Medicine (Baltimore) 2014;93:e296. [Crossref] [PubMed]
- Wu TH, Hsiue EH, Lee JH, et al. New data on clinical decisions in NSCLC patients with uncommon EGFR mutations. Expert Rev Respir Med 2017;11:51-5. [Crossref] [PubMed]
- Tu HY, Ke EE, Yang JJ, et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer 2017;114:96-102. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Pelosi G, Fabbri A, Papotti M, et al. Dissecting Pulmonary Large-Cell Carcinoma by Targeted Next Generation Sequencing of Several Cancer Genes Pushes Genotypic-Phenotypic Correlations to Emerge. J Thorac Oncol 2015;10:1560-9. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828-51. [Crossref] [PubMed]
- Janes MR, Zhang J, Li LS, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172:578-89.e17. [Crossref] [PubMed]
- Romero D. Two new agents target KRAS G12C. Nat Rev Clin Oncol 2020;17:6. [PubMed]
- Misale S, Fatherree JP, Cortez E, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25:796-807. [Crossref] [PubMed]
- Molina-Arcas M, Moore C, Rana S, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med 2019;11:eaaw7999. [Crossref] [PubMed]
- Wang H, Lv Q, Xu Y, et al. An integrative pharmacogenomics analysis identifies therapeutic targets in KRAS-mutant lung cancer. EBioMedicine 2019;49:106-17. [Crossref] [PubMed]
- Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913-21. [Crossref] [PubMed]
- Spoerke JM, O'Brien C, Huw L, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res 2012;18:6771-83. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther 2012;11:485-91. [Crossref] [PubMed]
- Gkolfinopoulos S, Mountzios G. Beyond EGFR and ALK: targeting rare mutations in advanced non-small cell lung cancer. Ann Transl Med 2018;6:142. [Crossref] [PubMed]
- Gao X, Zhu J, Chen L, et al. Clinical And Imageological Features Of Lung Squamous Cell Carcinoma With EGFR Mutations. Cancer Manag Res 2019;11:9017-24. [Crossref] [PubMed]


