
Gain of GAS5 reveals worse prognosis in kidney renal clear cell carcinoma and liver hepatocellular carcinoma from the Cancer Genome Atlas dataset
Introduction
Human cancer is a complex disease caused by the genetic and epigenetic alternations of two kinds of crucial genes: tumor suppressor genes and oncogenes (1). p53, one of the most famous tumor suppressor genes, controls DNA repair system and restrict chromosomal instability (2). While KRAS, as an essential oncogene, has been shown to promote cancer development and progression as well as influence the tumor microenvironment (3). However, some genes like TGF-β family members exert tumor suppressive effect early in tumorigenesis, but later they can lead to tumor progression and metastasis depending on signaling from growth factor receptors or activity of transcriptional regulators (4).
Being a long non-coding RNA and unable to encode protein, growth arrest-specific 5 (GAS5) has been reported to be a vital tumor suppressor and involved in apoptosis, proliferation, migration, invasion, cell cycle and metabolism of multiple cancers (5). Recent research reports that GAS5 suppressed the angiogenesis, invasion, and metastasis of colorectal cancer by inhibiting the activated Wnt/β‐catenin signaling pathway (6). In addition, GAS5 could inhibit cervical cancer cell proliferation, migration and invasion through miR-21 (7). Overexpression of GAS5 in ovarian cancer cells restrained proliferation, and was associated with inflammasome formation and pyroptosis (8).
However, some studies have revealed that GAS5 has a certain ability to promote cancer. Li et al. have revealed that GAS5 expression was increased in esophageal cancer cells and miR-301a was sponged to alleviate its pro-oncogene effects (9). Previous study showed that ectopic expressed GAS5 displayed an anti-apoptosis effect in liver cancer cell lines (10).
In this research, we analyzed the expression of GAS5 through data mining in The Cancer Genome Atlas (TCGA) datasets, and explored its prognostic value and potential regulatory mechanism in both kidney renal clear cell carcinoma (KIRC) and liver hepatocellular carcinoma (LIHC).
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2221).
Methods
Data from the TCGA dataset
Expression of GAS5 in most cancer types represented in the form of log2(TPM + 1) from the University of California Santa Cruz (UCSC) Xena Project were obtained from GEPIA (http://gepia.cancer-pku.cn/) (11), only those groups had more than 10 samples were included. And the correlation between GAS5 and ABHD2 as well as the survival data of patients (updated to June 2019) with KIRC and LIHC were downloaded from R2 website related to TCGA program (http://r2platform.com). DNA methylation data of GAS5 and abhydrolase domain containing 2 (ABHD2) (measured by the Infinium Human Methylation 450 BeadChip) in LIHC and KIRC were downloaded from the TCGA Wanderer (http://www.maplab.cat/wanderer) (12). All these data can be downloaded for free from the above public website. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
One-way ANOVA was used to detect differences for comparisons. Survival curves were generated by the Kaplan Scan by using median or scan as a cutoff in assessing whether GAS5 or ABHD2 has the potential to separate patient survival and test the P value in a log-rank test. Wilcoxon test was conducted to assess the DNA methylation of CpG sites of GAS5 or ABHD2 between normal and tumor sample in both KIRC and LIHC patients. P<0.05 was considered as statistically significant.
Results
Expression of GAS5 in most cancer types from TCGA program
To explore the expression of GAS5 in different kinds of cancers, we obtained RNA-sequencing data from TGCA in GEPIA (Figure 1). Nine types of tumor were excluded because they didn’t have enough normal tissue samples. By comparing the expression level of GAS5 between 14 kinds of cancer tissues and their normal tissues, we found that the expression of GAS5 had no significant difference in most of them, but was significantly upregulated in KIRC and LIHC (fold change >2, P<0.01).
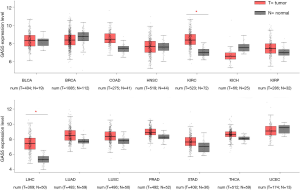
High GAS5 expression is associated with poor overall survival (OS) in both KIRC and LIHC
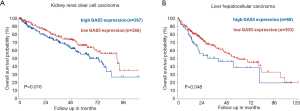
In order to validate its prognostic significance, we compared the association between GAS5 and OS using Kaplan-Meier survival curves (Figure 2). In KIRC patients, we separated them by the median expression of GAS5 into two groups, and the low GAS5 expression groups (n=266) had much longer OS than their respective high expression groups (n=267) (Figure 2A, P=0.016). In LIHC patients, we used scan as a cutoff to separated them into two groups, and the high GAS5 expression groups (n=68) had shorter OS than their low expression groups (n=303) (Figure 2B, P=0.048).
ABHD2 was highlighted between those genes co-related with GAS5 in both cancers
New biologically relevant hypotheses can be generated when we find out the related genes. Therefore, we performed HugoOnce mode analysis in R2 software to explore the relations of other genes with GAS5 in KIRC and LIHC. It summarized the results of a mini ontology analysis (Tables 1,2). In KIRC, those genes were involved with the process of DNA repair, apoptosis, cell cycle, kinase, membrane, signal transduction, transcription regulator activity and transcriptional repressor activity; while in LIHC, they were involved with DNA repair, development, membrane, transcription regulator activity and transcriptional repressor activity. Among them, membrane groups had the most significant P value. As a membrane protein, ABHD2 was highlighted in both KIRC and LIHC when we listed the top 20 genes (Tables 3,4).
Table 1
| Group | InSet | Total | % | P |
|---|---|---|---|---|
| All | 4,131 | 9,247 | 44.70 | 1 |
| DNA repair | 119 | 211 | 56.40 | 6.10E–04 |
| Transcription factor | 374 | 812 | 46.10 | 0.43 |
| Apoptosis | 315 | 633 | 49.80 | 0.01 |
| Cell cycle | 254 | 492 | 51.60 | 1.90E–03 |
| Development | 698 | 1,602 | 43.60 | 0.37 |
| Different | 299 | 682 | 43.80 | 0.66 |
| Drugtarget | 506 | 1,151 | 44.00 | 0.63 |
| Kinase | 334 | 649 | 51.50 | 5.00E–04 |
| Membrane | 2,025 | 4,889 | 41.40 | 4.70E–06 |
| Signal transduction | 1,258 | 3,026 | 41.60 | 6.00E–04 |
| Transcription regulator activity | 608 | 1,208 | 50.30 | 7.70E–05 |
| Transcriptional repressor activity | 98 | 180 | 54.40 | 8.40E–03 |
KIRC, kidney renal clear cell carcinoma.
Table 2
| Group | InSet | Total | % | P |
|---|---|---|---|---|
| All | 4,506 | 9,184 | 49.10 | 1 |
| DNA repair | 121 | 211 | 57.30 | 0.02 |
| Transcription factor | 394 | 811 | 48.60 | 0.78 |
| Apoptosis | 333 | 633 | 52.60 | 0.07 |
| Cell cycle | 263 | 492 | 53.50 | 0.05 |
| Development | 738 | 1,600 | 46.10 | 0.02 |
| Different | 318 | 681 | 46.70 | 0.22 |
| Drugtarget | 575 | 1,151 | 50.00 | 0.54 |
| Kinase | 342 | 649 | 52.70 | 0.06 |
| Membrane | 2,243 | 4,828 | 46.50 | 2.90E–04 |
| Signal transduction | 1,436 | 2,967 | 48.40 | 0.47 |
| Transcription regulator activity | 611 | 1,207 | 50.60 | 0.28 |
| Transcriptional repressor activity | 103 | 180 | 57.20 | 0.03 |
LIHC, liver hepatocellular carcinoma.
Table 3
| HUGO | Function | R | P | HUGO | Function | R | P |
|---|---|---|---|---|---|---|---|
| ZFAS1 | – | 0.791 | 2.2E–113 | SNX19 | Signal | –0.576 | 1.5E–46 |
| RPL5 | – | 0.784 | 5.5E–110 | AP1G1 | Membrane | –0.536 | 3.8E–39 |
| RPS8 | – | 0.765 | 1.2E–101 | FICD | – | –0.518 | 3.1E–36 |
| RPS18 | – | 0.749 | 6E–95 | USP9X | – | –0.51 | 6E–35 |
| EPB41L4A-AS1 | – | 0.738 | 8.1E–91 | PEX26 | Membrane | –0.507 | 2.5E–34 |
| RPS3 | – | 0.731 | 3.4E–88 | GLYR1 | – | –0.505 | 4.9E–34 |
| RPL31 | – | 0.72 | 3.2E–84 | AAK1 | Kinase | –0.505 | 4.2E–34 |
| RPL12 | – | 0.714 | 3.7E–82 | ADAT1 | – | –0.504 | 6.1E–34 |
| RPL18 | – | 0.712 | 9.6E–82 | ANKFY1 | Membrane | –0.502 | 1.4E–33 |
| RPL11 | – | 0.708 | 3.8E–80 | ABHD2 | Membrane | –0.501 | 2E–33 |
| RPS6 | – | 0.699 | 1.9E–77 | KIAA0232 | – | –0.498 | 5.8E–33 |
| RPS24 | – | 0.688 | 5.6E–74 | UGGT1 | – | –0.489 | 1.2E–31 |
| RPL34 | – | 0.687 | 8.5E–74 | ITPR2 | Membrane | –0.488 | 1.6E–31 |
| RPL4 | – | 0.684 | 8.2E–73 | NDUFS1 | Apoptosis | –0.484 | 7.1E–31 |
| RPL38 | – | 0.684 | 8.9E–73 | DYNLL2 | – | –0.484 | 5.6E–31 |
| RPL10 | – | 0.678 | 4.1E–71 | NSF | – | –0.48 | 2.4E–30 |
| LRRC75A-AS1 | – | 0.678 | 6.1E–71 | GSK3B | Apoptosis | –0.48 | 2.3E–30 |
| RPL10A | – | 0.677 | 7.9E–71 | SMC1A | DNArepair | –0.476 | 9.5E–30 |
| RPL30 | – | 0.676 | 1.7E–70 | ATP10D | DNArepair | –0.476 | 8.1E–30 |
| RPL19 | – | 0.675 | 2.80E–70 | DIRC2 | Membrane | –0.476 | 8.3E–30 |
‘–’: The function of these proteins were not mentioned in the R2 websites. GAS5, growth arrest-specific 5; KIRC, kidney renal clear cell carcinoma.
Table 4
| HUGO | Function | R | P | HUGO | Function | R | P |
|---|---|---|---|---|---|---|---|
| ZFAS1 | – | 0.792 | 3.8E–79 | RSC1A1 | – | –0.647 | 1.2E–43 |
| RPS24 | – | 0.764 | 1.9E–70 | SCP2 | – | –0.646 | 1.4E–43 |
| RPS3 | – | 0.764 | 2.2E–70 | AP5M1 | – | –0.642 | 8.1E–43 |
| RPS18 | – | 0.762 | 9.9E–70 | LONP2 | – | –0.634 | 2E–41 |
| RPL31 | – | 0.752 | 5.2E–67 | DDI2 | – | –0.632 | 4.8E–41 |
| RPL6 | – | 0.752 | 7.4E–67 | ATP11C | Membrane | –0.626 | 3.8E–40 |
| SNHG6 | – | 0.75 | 2.4E–66 | WDR7 | – | –0.622 | 1.9E–39 |
| RPL14 | – | 0.748 | 5.1E–66 | SLC41A2 | – | –0.622 | 2.3E–39 |
| RPL8 | – | 0.746 | 1.8E–65 | ERCC4 | DNArepair | –0.618 | 7.6E–39 |
| RPL23 | – | 0.74 | 1.1E–63 | ALDH6A1 | Drugtarget | –0.61 | 1.6E–37 |
| RPL37A | – | 0.739 | 1.4E–63 | SLC35A3 | Membrane | –0.609 | 2.2E–37 |
| RPLP0 | – | 0.738 | 2.3E–63 | SLC30A4 | Membrane | –0.602 | 2.3E–36 |
| RPS10 | – | 0.738 | 2.7E–63 | MYLK | Kinase | –0.601 | 3.5E–36 |
| RPL32 | – | 0.736 | 8.9E–63 | APOOL | – | –0.594 | 3.7E–35 |
| RPS7 | – | 0.733 | 4.7E–62 | ZYG11B | – | –0.593 | 6.4E–35 |
| EPB41L4A-AS1 | – | 0.732 | 8.7E–62 | CPED1 | – | –0.592 | 8.6E–35 |
| GNB2L1 | Signal | 0.731 | 2.2E–61 | EFCAB14 | – | –0.588 | 3.2E–34 |
| RPSA | Membrane | 0.729 | 7.9E–61 | HERC3 | – | –0.586 | 6.2E–34 |
| RPL37 | – | 0.725 | 6.2E–60 | ABHD2 | Membrane | –0.584 | 1.3E–33 |
| TAF1D | – | 0.725 | 7.7E–60 | AGL | – | –0.583 | 1.7E–33 |
‘–’: The function of these proteins were not mentioned in the R2 websites. GAS5, growth arrest-specific 5; LIHC, liver hepatocellular carcinoma.
ABHD2 is negatively correlated with GAS5
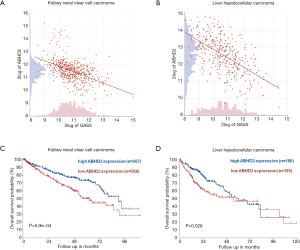
Data mining in R2 confirmed a moderately negative correlation between the expression of ABHD2 and GAS5 in both KIRC (R=–0.501, P<0.001) and LIHC (R=–0.584, P<0.001) cancers (Figure 3A,B). Also, we wanted to verify the association of ABHD2 with OS of those cancer patients. Thus, the low ABHD2 expression groups had significantly shorter OS than their respective high expression groups in KIRC (Figure 3C, P=0.00059) and LIHC (Figure 3D, P=0.026) patients.
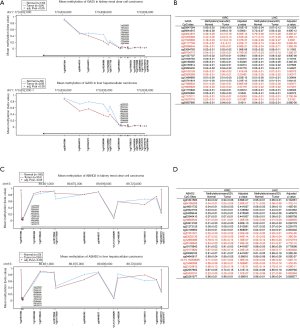
DNA methylation might regulate the transcription of GAS5 and ABHD2 in both cancers
To further examine the potential regulatory effect of DNA methylation on GAS5 and ABHD2 expression, we compared their status in the TCGA Wanderer. Figure 4A,B showed that 12 CpG sites of GAS5 (cg03044573, cg16290996, cg17025683, cg07177756, cg05396044, cg06704773, cg04102126, cg02955369, cg17868533, cg16905926, cg17170111, cg10578779) were significantly hypermethylated in the normal tissues compared with tumor tissues. In comparison, Figure 4C,D showed that 11 CpG sites of ABHD2 (cg04788288, cg16663533, cg27351321, cg03025135, cg12850396, cg25156843, cg10173620, ch.15.1525362R, cg13460556, cg23124613, cg03432151) were significantly hypomethylated in the normal tissues compared with tumor tissues. These results inferred that DNA methylation might regulate their transcriptions in both cancers.
Discussion
Published data showed that the expression of GAS5 was decreased in renal cell carcinoma cell lines (13). Similarly, reduced GAS5 expression was detected in clear cell renal cell carcinoma tissues and independently correlated with poor disease free survival of patients; their data revealed that GAS5 may function as a competing endogenous RNA (ceRNA) to positively regulate hZIP1 by sponging miR-223 (14). Latest research reported GAS5 downregulation promoted responsive renal cell carcinoma cells resistant to sorafenib treatment through miR-21/SOX5 axis (15). However, our research demonstrated that the mRNA level of GAS5 was significantly increased more than two folds in TCGA-KIRC dataset, probably because TCGA-KIRC dataset had much more samples than those previous studies. Moreover, high GAS5 expression was associated with poor OS, which predicted that GAS5 could be a prognostic marker for KIRC patients.
There are contrary results about GAS5 association with liver cancer. Two previous studies reported that downregulation of GAS5 was detected in liver cancer tissues compared with adjacent tissues and its reduction was associated with poor OS (16,17). Some other studies revealed that GAS5 could suppress metastasis via miR-21 (18) and inhibit the epithelial mesenchymal transition (EMT) of liver cancer cells (19). In contrary, knockdown of GAS5 partially revert the anti-apoptosis effect in liver cancer cell lines (10). A bioinformatic analysis demonstrated that high GAS5 was an independent predictor of shorter relapse free survival rather than OS in TCGA-LIHC (20). In the contrary, when we used scan as a cutoff to separated LIHC patients into two groups, results showed that high GAS5 level was associated with poor OS.
Therefore, we speculated that GAS5 might have its unique role and exert oncogenic effect in the development and progression of KIRC and LIHC. And, GAS5 might have different functions depending on different genetic backgrounds. In order to find out its new mechanism, we explored the correlated genes of GAS5. Results showed that GAS5 might be involved with DNA repair, development, apoptosis, cell cycle, kinase, membrane, signal transduction, transcription regulator activity and transcriptional repressor activity. Among them, ABHD2 was found to be negatively correlated with GAS5, and its low level could predict poor OS in KIRC and LIHC patients, which has never been reported before.
As a member of the α/β hydrolase superfamily, ABHD2 was reported to possess triacylglycerol (TAG) lipase along with ester hydrolysing capacity and regulate the lipid metabolism and signal transduction (21). ABHD2 deficient in aged mice led to reduction of alveolar type II cells and accumulation of macrophages in the lungs; in addition, ABHD2 could affect macrophage recruitment to atherosclerotic lesions. Thus, ABHD2 seemed to play a significant role in chronic diseases like emphysema and atherosclerosis (22,23). A study showed that knockdown of ABHD2 blocks hepatitis B virus propagation (24); while others reported that ABHD2 knockdown could reduce cell death by attenuating calcium release from the endoplasmic reticulum and mitochondrial calcium uptake in primary mouse lung fibroblasts stimulated with A23187 (25).
There are two reports of correlation between ABHD2 and human cancer. In prostate cancer, ABHD2 was thought to promote cancer cell proliferation and migration (26); while suppression of ABHD2 led to chemoresistance and poor prognosis in ovarian cancer patients (27). Although there were no clues about how ABHD2 and GAS5 involved in their mutual regulation network of KIRC and LIHC, interestingly, our data implied that high GAS5 expression had lower DNA methylation status in tumor tissues compared to normal tissues in both KIRC and LIHC patients; while low ABHD2 expression had higher DNA methylation status compared to normal tissues in both KIRC and LIHC patients. Therefore, DNA methylation might regulate their transcriptions in both cancers.
To our knowledge, our research is the first to report that GAS5 might be involved with ABHD2 in KIRC and LIHC. Although the detailed molecular mechanisms remain unclear and need further verification, our findings demonstrated GAS5 as a prognostic marker for KIRC and LIHC, and could be useful for the development of biomarkers for those cancer treatments.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2221
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2221). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang LH, Wu CF, Rajasekaran N, et al. Loss of tumor suppressor gene function in human cancer an overview. Cell Physiol Biochem 2018;51:2647-93. [Crossref] [PubMed]
- Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062-78. [Crossref] [PubMed]
- Aguirre AJ, Hahn WC. Synthetic lethal vulnerabilities in KRAS-mutant cancers. Cold Spring Harb Perspect Med 2018;8:a031518. [Crossref] [PubMed]
- Nyati S, Schinske-Sebolt K, Pitchiaya S, et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-beta signaling. Sci Signal 2015;8:ra1. [Crossref] [PubMed]
- Lu S, Su Z, Fu W, et al. Altered expression of long non-coding RNA GAS5 in digestive tumors. Biosci Rep 2019;39:BSR20180789. [Crossref] [PubMed]
- Song J, Shu H, Zhang L, et al. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wntbeta-catenin signaling pathway. J Cell Biochem 2019; [Epub ahead of print].
- Wen Q, Liu Y, Lyu H, et al. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer 2017;27:1096-108. [Crossref] [PubMed]
- Li J, Yang C, Li Y, et al. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep 2018;38:BSR20171150. [Crossref] [PubMed]
- Li W, Zhao W, Lu Z, et al. Long noncoding RNA GAS5 promotes proliferation, migration, and invasion by regulation of miR-301a in esophageal cancer. Oncol Res 2018;26:1285-94. [Crossref] [PubMed]
- Tao R, Hu S, Wang S, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis 2015;36:1136-43. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Díez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015;8:22. [Crossref] [PubMed]
- Qiao HP, Gao WS, Huo JX, et al. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev 2013;14:1077-82. [Crossref] [PubMed]
- Dong X, Kong C, Liu X, et al. GAS5 functions as a ceRNA to regulate hZIP1 expression by sponging miR-223 in clear cell renal cell carcinoma. Am J Cancer Res 2018;8:1414-26. [PubMed]
- Liu L, Pang X, Shang W, et al. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle 2019;18:257-63. [Crossref] [PubMed]
- Tu ZQ, Li RJ, Mei JZ, et al. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol 2014;7:4303-9. [PubMed]
- Chang L, Li C, Lan T, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep 2016;13:1541-50. [Crossref] [PubMed]
- Hu L, Ye H, Huang G, et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol 2016;37:2691-702. [Crossref] [PubMed]
- Chen CY, Chen CC, Shieh TM, et al. Corylin suppresses hepatocellular carcinoma progression via the inhibition of epithelial-mesenchymal transition, mediated by long noncoding RNA GAS5. Int J Mol Sci 2018;19:380. [Crossref] [PubMed]
- Zhu Q, Yang H, Cheng P, et al. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. Biofactors 2019;45:244-52. [Crossref] [PubMed]
- Naresh Kumar M, Thunuguntla VBSC, Veeramachaneni GK, et al. Molecular characterization of human ABHD2 as TAG lipase and ester hydrolase. Biosci Rep 2016;36:e00358. [Crossref] [PubMed]
- Miyata K, Nakayama M, Mizuta S, et al. Elevated mature macrophage expression of human ABHD2 gene in vulnerable plaque. Biochem Biophys Res Commun 2008;365:207-13. [Crossref] [PubMed]
- Jin S, Zhao G, Li Z, et al. Age-related pulmonary emphysema in mice lacking alpha/beta hydrolase domain containing 2 gene. Biochem Biophys Res Commun 2009;380:419-24. [Crossref] [PubMed]
- Ding X, Yang J, Wang S. Antisense oligonucleotides targeting abhydrolase domain containing 2 block human hepatitis B virus propagation. Oligonucleotides 2011;21:77-84. [Crossref] [PubMed]
- Yun B, Lee H, Powell R, et al. Regulation of calcium release from the endoplasmic reticulum by the serine hydrolase ABHD2. Biochem Biophys Res Commun 2017;490:1226-31. [Crossref] [PubMed]
- Obinata D, Takada S, Takayama K, et al. Abhydrolase domain containing 2, an androgen target gene, promotes prostate cancer cell proliferation and migration. Eur J Cancer 2016;57:39-49. [Crossref] [PubMed]
- Yamanoi K, Matsumura N, Murphy SK, et al. Suppression of ABHD2, identified through a functional genomics screen, causes anoikis resistance, chemoresistance and poor prognosis in ovarian cancer. Oncotarget 2016;7:47620-36. [Crossref] [PubMed]


