
Identification of potential biomarkers and available drugs for oral squamous cell carcinoma
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of oral tumor, accounting for 90% of total oral tumor cases. The annual global number of new cases is approximately 650,000 (1), placing OSCC eighth for incidence among all malignant tumors. Hasegawa et al. Have reported an increasing trend for a younger age of onset among OSCC patients (2). In recent years, the number of cases has increased, and the 5-year overall survival rate has decreased from less than 50% to less than 30%. These statistics provide a stark reminder that research into the prevention, diagnosis and treatment of OSCC needs to be intensified. The low survival rate of OSCC is mainly due to cervical lymph node metastasis at the time of diagnosis, and the prognosis for this group of patients is often poor (3). Xiao et al. Indicated that the expression of Bit-1 messenger RNA (mRNA) and its protein, which is related to the occurrence, metastasis, and invasion of OSCC, could be a key factor in the disease, and may be a promising diagnostic and therapeutic target in the treatment of OSCC (4). However, despite an extensive exploration of OSCC at the molecular level, no optimal target has been identified to date.
With the development of biological research, a large amount of mmics data has been obtained through the application of high-throughput experimental methods. Besides, the application of microarray analysis to transcriptional research has offered us a deeper understanding of the expressive landscape of the biological mechanisms of multifactorial disease (5). The integration of multiple microarray datasets has also generated disease-associated mRNA profiles for cancer screening. The experimental conditions of each data set are clinically and technically different, but it remains to be seen that whether the common differentially expressed genes (DEGs) associated with OSCC in multiple data sets can provide new perspectives for identifying key genes as potential targets for diagnosis and drug targeting.
Currently, increased attention is being paid to the sharing and integration of omics data for the study of multi-factorial disease mechanisms. To further promote the sharing of data, a proposal has been made to register biological experimental data in a public database. This utilization of pooled microarray gene expression data sets is an excellent method that can reduce hybridization costs and also compensate for insufficient amounts of mRNA sampling (6). To promote biomedical research, the National Biotechnology Information Center has developed the Gene Expression Omnibus (GEO), which is a publicly available integrated database for the collection and sharing of transcriptome data (7-9). ArrayExpress is another public database for high-throughput functional genomic data that comprises two parts: the ArrayExpress Repository, which offers the minimum information about a microarray experiments, and the ArrayExpress Data Warehouse, a database of gene expression profiles selected from a repository that is consistently reannotated (10,11). At present, many scholars are using microarray analysis to explore the pathogenesis of periodontitis (12,13).
In this study, we used comprehensive bioinformatics analyses to uncover the potential genes and signaling pathways in OSCC. First, we selected and analyzed two pooled microarray platform datasets in the GEO database. Then, the DEGs were identified between the OSCC and normal groups. Next, Gene Ontology (GO), signaling pathway enrichment annotation, and protein–protein interaction (PPI) network analyses were performed among these dysregulated genes through the application of different bioinformatics methods (14). This allowed us to identify the potential gene biomarkers and correlated pathways, that might be associated with OSCC and which may provide possible biological targets for the therapeutic treatment of OSCC. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2500).
Methods
Microarray data
GEO (http://www.ncbi.nlm.nih.gov/geo) is a database repository, storing high-throughput gene expression data and hybridization arrays, chips, and microarrays. We downloaded two gene expression datasets from GEO: GSE30784 and GSE23558. The platforms that they were based on were GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array) and GPL6480 (Whole Human Genome Microarray 4x44K G4112F). GSE30784 contained 167 OSCC and 45 normal oral tissue samples. GSE23558 comprised 27 OSCC and 4 normal samples (Table 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Series | Platform | Affymetrix gene chip | Samples |
|---|---|---|---|
| GSE23558 | GPL6480 | Whole Human Genome Microarray 4×44K G4112F | 31 |
| GSE30784 | GPL570 | (Affymetrix Human Genome U133 Plus 2.0 Array | 212 |
Data processing of DEGs
DEGs between the OSCC and normal specimens were identified using the GEO2R online tools (http://www.ncbi.nlm.nih.gov/geo/geo2r/) with the criteria set as |logFC| >2 and an adjusted P value <0.05. Then, the raw data in TXT format were checked with Venn software (http://bioinformatics.psb.ugent.be/webtools/Venn/) to detect the common DEGs between the two data sets. The DEGs with a log FC <−2 were considered as a downregulated gene, while the DEGs with a log FC >2 were considered as upregulated genes.
GO and pathway enrichment analysis
GO analysis is the most commonly used method for defining genes. With this method, RNA or protein products are analyzed to identify unique biological characteristics of high-throughput transcriptome or genomic data. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a collection of databases dealing with genomes, diseases, biological pathways, drugs, and chemical materials (14). The Database for Annotation, Visualization and Integrated Discovery (DAVID) is an online bioinformatics tool that identifies the function of a large number of genes or proteins (6). We used DAVID to visualize the DEG enrichment of biological processes (BP), molecular functions (MF), and cellular components (CC) (FDR <0.05).
PPI network construction and module analysis
We used the online Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org) to identify the interactive relationships of the overlapping DEGs (6). A confidence score ≥0.4 was considered to be significant. The Cytoscape software (http://www.cytoscape.org) was used to construct and visualize a PPI network of common DEGs (14). The plugin cytoHubba was used to select the top 10 hub genes from the PPI network (score >6) (15).
Drug-gene interaction and the functional analysis of potential genes
We explored the interaction between genes and drugs in order to obtain the potential application of new treatment indications for OSCC. The Drug-Gene Interaction Database (DGIdb: https://www.dgidb.org) is an open-source database that supports searching, browsing, and filtering of information on drug-gene interactions from more than 30 trusted sources (16). The module genes, as the potential targets, were pasted into the DGIdb to search for existing drugs or compounds. The genes that had matched drugs were obtained, and a functional enrichment analysis was performed.
Statistical analysis
A moderated t-test was applied to identify DEGs, and Fisher's exact test was used to test for enrichment in GO terms and KEGG pathways.
Results
The identification of DEGs in OSCC
A total of 1628 DEGs (610 in GSE30784 and 1018 in GSE23558) were identified by GEO2R. There were 199 genes that overlapped between the two data sets, as shown in the Venn diagram generated by FunRich (17). Through Venn analysis, the genes that were consistently upregulated and downregulated were screened out. As a result, we found 198 upregulated DEGs (Figure 1A) and 1 downregulated DEG (Figure 1B) (Table 2).
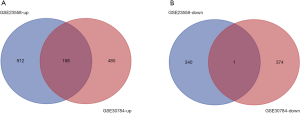
Table 2
| DEGs | Gene name |
|---|---|
| Upregulated | TMPRSS11B, PTN, GSTA1, CHRDL1, EHF, CILP, SLC6A4, UPK1A, CAPNS2, SYNPO2, SFTA2, CXCL13, ENDOU, AADAC, EPHX3, MPP7, LOC100506098, KRT13, ANKRD35, A2ML1, KIAA1211L, SORT1, SCIN, TSPAN8, TMPRSS11E, CAPN14, PAX9, TNNC2, GALNT5, FUT6, LAMB4, MYOT, MAL, VSIG10L, NEBL, ATP13A4, C15orf62, TP53INP2, HLF, CLDN7, SNX31, ANXA9, MUC15, SPINK5, EMP1, TM7SF2, PPL, ABCA8, VIT, CYP3A5, SYTL4, PITX1, IGHM, KRT24, NDRG2, MEIS1, ADH7, FAM3B, ARHGEF26, IGLJ3, FCER1A, DCT, RBM20, TYRP1, CD207, LOC105379426, SH3BGRL2, ANKRD20A5P, MYH11, TGM3, GALNT12, CYP2E1, ABI3BP, SPINK7, CLDN10, CYP4F12, SFRP4, LRRC15, SCNN1B, GRHL3, MUC20, PRSS27, RBP7, KLK12, PRSS3, ACPP, HPGD, CWH43, SCEL, BNIPL, HRASLS, SAMD5, TMPRSS2, EPHX2, ANGPTL1, CXCL17, LOC441178, TMEM100, ATP6V0A4, ASPN SERPINB11, GREM2, DPT, SLC27A6, MFAP4, TMEM45B, ALOX12, IKZF2, TPRG1, MGLL, SASH1, OGN, SCARA5, HCG22, MAOB, CLCA4, ATP1A2, IGSF10, CLIC3, KLF8, SLC16A6, KRT4, IGKC, KLK13, LIFR, SPNS2, EXPH5, C2orf54, CYP2C18, ALDH3A1, CEACAM5, GDPD3, FUT3, MEOX2, IL36A, APOD, COMP, DIO2, PPM1L, CRNN, MACC1, SUSD4, CEACAM7, HBB, IL1RN, FAM189A2, PAIP2B, CA3 KRT19, FMO2, ANKRD20A11P, DEPTOR, MYZAP, ATP6V1C2, COBL, TGFBR3, MYRIP, FAM221A, SCNN1A, CAB39L, CRTAC1, FAM107A, NMU, PGM5, MANSC1, GPX3, TCP11L2, PPP1R3C, RRAGD, SLURP1, TCEA3, MMRN1, CEACAM1, PYGM, PLP1, RAET1E, PSCA, MAMDC2, TTC9, FAM3D, SLC4A4, CYP2J2, TFF3, C2orf40, DAPL1, ATP10B, SELENBP1, LNX1, GULP1, CFD, SYNGR1, CYP4B1, EYA2, ST6GALNAC1 HOPX, BICDL2, CRCT1 |
| Downregulated | AGR2 |
GO and pathway enrichment analysis
The functional annotation of genes was visualized using the DAVID website, and the GO and signaling pathway enrichment of DEGs was visualized. Figure 2 and Table 3 show the significantly enriched BP, CC, and MF terms for the DEGs. In the BP annotations, DEGs were mainly involved in the “epoxygenase P450 pathway” and the “xenobiotic metabolic process”. In the CC annotations, DEGs were significantly involved in the “organelle membrane”, “extracellular space”, “extracellular region”, and “extracellular exosome”. In the MF annotations, DEGs were evident in “serine-type peptidase activity”, and “monooxygenase activity”. Of the KEGG pathways, the DEGs were enriched in “drug metabolism - cytochrome P450”.
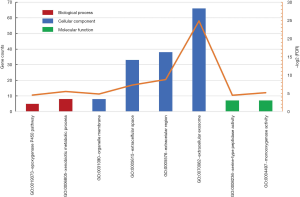
Table 3
| Term | Category | Description | FDR | Count |
|---|---|---|---|---|
| GO:0019373 | BP | Epoxygenase P450 pathway | 0.043608 | 5 |
| GO:0006805 | BP | Xenobiotic metabolic process | 0.02217 | 8 |
| GO:0031090 | CC | Organelle membrane | 0.035542 | 8 |
| GO:0005615 | CC | Extracellular space | 0.006491 | 33 |
| GO:0005576 | CC | Extracellular region | 0.002268 | 38 |
| GO:0070062 | CC | Extracellular exosome | 3.26E-08 | 66 |
| GO:0008236 | MF | Serine-type peptidase activity | 0.045136 | 7 |
| GO:0004497 | MF | Monooxygenase activity | 0.027988 | 7 |
| hsa00982 | KEGG | Drug metabolism cytochrome P450 | 0.018393 | 7 |
PPI network and module analysis
The STRING App in the Cytoscape software was used to analyze 199 DEGs that had been entered into the STRING database. A total of 125 genes/nodes with 148 edges were enriched in the construction of the PPI network, while 67 genes did not fall into the network (Figure 3). Furthermore, a significant gene module was selected to cluster all genes using the cytoHubba app built in Cytoscape. As shown in Figure 4, the top four hub genes were CYP2E1, SCEL, KRT4, and KRT19 (score >6). All parameters in cytoHubba were set by degree. A drug-gene interaction analysis was performed on the genes or nodes in the gene modules.
Drug-gene interaction and functional analysis of potential genes
The 10 potential genes clustered in the significant gene module were selected for drug-gene interaction analysis. In humans, one existing drug, etoposide, was shown to target the CYP2E1 gene.
The top 10 hub genes clustered in the significant gene module were uploaded in QuartataWeb (http://quartata.csb.pitt.edu) for drug-gene interaction analysis. The results, including genes, predicted drugs, and pathways with the criteriom of P<0.05 were processed using the Sankey package (https://cran.r-project.org/web/packages/sankeywheel/index.html) built in R software (version 3.5.2). As shown in Figure 5, only three genes (CYP2E1, CYP2J2, and GSTA1) were matched to the predicted drugs, and the already-applied drug etoposide was the inhibitor of CYP2E1. In the KEGG pathway, DEGs were enriched in several processes including “metabolism of xenobiotics by cytochrome P450”, “drug metabolism - other enzymes”, and “linoleic acid metabolism”.
Discussion
At present, the tumor markers commonly used in the diagnosis of OSCC have been criticized for a lack of sensitivity and specificity. Diagnosis of OSCC is mainly based on imaging methods, which is an inappropriate strategy for patients suffering in the late stages of a malignant illness (2). Common treatment methods include radiotherapy, chemotherapy, and traditional surgery, but these are often traumatic and can significantly affect patient wellbeing and quality of life (18). According to Bloebaum et al. (19), approximately 20% of OSCC patients treated with traditional surgical procedures experience recurrence. At present, radiotherapy and chemotherapy are also commonly used to treat cancer. However, they render the immune system more vulnerable, thus making the patient susceptible to other diseases during their recovery from treatment (20). To date, molecular targeted therapy has not been used in OSCC, so our intention in this study was to expand on previous findings of OSCC-relevant genes, in order to assist in the clinical diagnosis and treatment of OSCC in the future.
In this study, several important components in bioinformatics, which combines computer science with biological science, were employed to make the exploration of DEGs more concise. Public databases were also used in this study for the purpose of improving efficiency.
Two microarrays related to OSCC tissues were obtained from the GEO database. Using GEO2R, 610 and 1018 DEGs were screened out from GSE30784 and GSE23558, respectively, and there were 199 common DEGs between the two microarrays. Subsequent GO enrichment analysis showed that the DEGs in BP were mainly distributed in the “epoxygenase P450 pathway” and the “xenobiotic metabolic process”. In the CC annotations, DEGs were significantly involved in the “organelle membrane”, “extracellular space”, “extracellular region”, and “extracellular exosome”. In the MF annotations, DEGs were significantly involved in “serine-type peptidase activity” and “monooxygenase activity”. In the KEGG pathway, DEGs were enriched in “drug metabolism - cytochrome P450”. These finding suggest that the occurrence and development of OSCC might be related to cellular matrix protein variations and the drug metabolic pathway. Through comprehensive analysis of gene expression profiles, we have been able to identify potential molecular biomarkers (CYP2E1, SCEL, KRT4, and KRT19) and new indications for existing drugs (etoposide).
In this study, we found that proteins such as “SCEL”, “KRT4”, and “KRT19” all had strong interactions, suggesting that these proteins may have a greater impact on the occurrence and development of OSCC, and indicating that they could be expected to become good markers for the diagnosis and treatment of OSCC in the future. Sciellin (SCEL) is a precursor of the cornified envelope, which is a protein enriched in the intima of arteries. SCEL is closely related to the stress characteristics of stratified epithelium (21). SCEL contains 16 inexact repeats of 20 amino acids and a LIM domain (derived from LIN-11, Isl1, and MEC-3) that may function as a protein interaction module to regulate the localization of SCEL and protein assembly in the cornified envelope (22). Based on the molecular characteristics of SCEL, we hypothesize that it may be closely related to the metastasis of cancer cells. Researches has shown that SCEL knockdown increased colorectal cancer cell migration and invasion, while its overexpression had the opposite effect. SCEL can activate mesenchymal-to-epithelial transition (MET) through a SCEL-β-catenin-E-cadherin axis (23), which raises the possibility that the lymph node metastasis of OSCC might be suppressed via intentional upregulation of the SCEL level. KRT4 and KRT19, both members of the keratin gene family, are major proteins found in the epidermis and hair follicles. As intermediate filament proteins, these two critical agents play several important roles within the cell. OSCC is a lesion of the oral epithelium. We speculated that the variation of KRT4 and KRT19 might promote the development of OSCC. A previous study found that KRT4 gene mutation was the molecular basis of oral white sponge nevus (WSN). In some cases, WSN is characterized by eosinophilic concentration of the area around the nucleus in the superficial epithelium. Other white keratosis, such as the precancerous condition lichen planus, may also have similar microscopic results (24). KRT19-positive hepatocellular carcinoma (HCC) is well known to have a higher malignant potential than K19-negative HCC. Studies have shown that KRT19 can reduce the expression of E-cadherin to enhance tumor invasion, and can also inhibit the senescence and apoptosis of HCC cells, improving their survival rate (25). Accordingly, this information indicates that KRT19 activation may promote important physiological processes in OSCC, including cancer cell survival, invasion, and angiogenesis.
Cytochrome P450 family 2 subfamily E member 1 (CYP2E1) was found to be the gene with significant difference between the two microarrays. Human cytochrome P450 (CYP) is a phase I enzyme which plays an important role in the metabolic activation of numerous procarcinogens (26). CYP2E1 represents a major CYP isoform. In the main, this gene is constitutively expressed in the liver but is also found to a lesser extent in other organs and tissues, including human urothelial cells. The extracellular ethanol-DNA adducts were reported to be significantly increased in HepG2 cells overexpressing CYP2E1 after exposure to ethanol (27), and this has also been seen in some patients with alcoholic fatty liver disease and fibrosis (28). In the presence of a specific CYP2E1 inhibitor, clomethiazole (CMZ), these DNA adducts can be significantly reduced (27,29). Furthermore, the level of this outer ring ethanol-DNA adduct can also be observed in esophageal biopsy specimens from human patients with alcoholic esophageal cancer (30). There have been few studies on the relationship between CYP2E1 and OSCC. In this study, CYP2E1 was the gene with the highest DEG score. Combined with the results of previous studies, it is reasonable to state that the overexpression of CYP2E1 may play a key role in the occurrence and development of OSCC. Etoposide, the inhibitor of CYP2E1, is an antitumor drug currently used in the treatment of small cell lung cancer (31), testicular cancer (32), and lymphoma (33). Since the introduction of etoposide in 1971, chemical and biological researchers have conducted substantial research on its mechanism of action and its powerful anti-tumor activity. This drug acts by stabilizing a normally transient DNA-topoisomerase II complex, thus increasing the concentration of double-stranded DNA breaks. This phenomenon triggers mutagenic and cell death pathways. The function of topoisomerase II is understood in some detail, as is the mechanism of inhibition of etoposide at a molecular level (34). Etoposide has some shortcomings, including limited neoplastic activity against several solid tumors, including non-small cell lung cancer, cross-resistance to multidrug-resistant tumor cell lines, and low bioavailability (35). The use of etoposide in clinical practice raises the possibility that this drug could play a currently unknown role in the treatment and prognosis of OSCC by inhibiting the expression of CYP2E1.
To date, there has been little research into the genes and drugs that might be implicated in the treatment of OSCC. Although we used comprehensive analyses to identify genes and existing drugs through the GEO datasets, experiments conducted at the molecular level are eagerly awaited to support our results. We hope that the results of this experiment can enrich the study of potential biomarkers and available drug treatments for OSCC, and assist in the clinical diagnosis and targeted treatment of this malignancy in the future.
Conclusions
By applying a series of bioinformatics methods to gene expression profiling, we acquired four potential biomarkers (CYP2E1, SCEL, KRT4, and KRT19) and one existing drug (etoposide) for OSCC. These four markers are closely related to the occurrence, development, and prognosis of tumors, among which SCEL may play a role in the migration of cancer cells by promoting mesenchymal transformation of the epithelium. KRT4 and some oral cancer precursors are closely related. KRT19 directly promotes cancer cell survival, invasion, and angiogenesis, and CYP2E1 plays an important role in the activation of various carcinogenic substances. Also, etoposide, an anti-tumor drug, that is already in clinical use, inhibits the expression of CYP2E1. We hope that the results of this study will provide insight for new study targets and drug indications for OSCC.
Acknowledgments
Funding: This work is supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2500
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form. The authors have no conflicts of interest to declare (available at http://dx.doi.org/10.21037/tcr-20-2500).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lin SH, Wang HK, Yeh KT, et al. c-MYC expression in T (III/IV) stage oral squamous cell carcinoma (OSCC) patients. Cancer Manag Res 2019;11:5163-9. [Crossref] [PubMed]
- Hasegawa T, Yanamoto S, Otsuru M, et al. Retrospective study of treatment outcomes after postoperative chemoradiotherapy in Japanese oral squamous cell carcinoma patients with risk factors of recurrence. Oral Surg Oral Med Oral Pathol Oral Radiol 2017;123:524-30. [Crossref] [PubMed]
- Khanna V, Karjodkar F, Robbins S, et al. Estimation of serum ferritin level in potentially malignant disorders, oral squamous cell carcinoma, and treated cases of oral squamous cell carcinoma. J Cancer Res Ther 2017;13:550-5. [PubMed]
- Xiao Y, Yang Y, Wang J, et al. Bit1 Regulates Cell Migration and Survival in Oral Squamous Cell Carcinoma. J Oral Maxillofac Surg 2017. [Epub ahead of pring]. doi:
10.1016/j.joms.2017.03.027 . - Zhai J, Qi A, Zhang Y, et al. Bioinformatics Analysis for Multiple Gene Expression Profiles in Sepsis. Med Sci Monit 2020;26:e920818. [Crossref] [PubMed]
- Xu Z, Zhou Y, Cao Y, et al. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol 2016;33:130. [Crossref] [PubMed]
- Dallavilla T, Bertelli M, Morresi A, et al. Bioinformatic analysis indicates that SARS-CoV-2 is unrelated to known artificial coronaviruses. Eur Rev Med Pharmacol Sci 2020;24:4558-64. [PubMed]
- Harada S, Akita H, Tsubaki M, et al. Dual graph convolutional neural network for predicting chemical networks. BMC Bioinformatics 2020;21:94. [Crossref] [PubMed]
- Xu X, Qiu B, Yi P, et al. Overexpression of miR-206 in osteosarcoma and its associated molecular mechanisms as assessed through TCGA and GEO databases. Oncol Lett 2020;19:1751-8. [Crossref] [PubMed]
- Patra BG, Maroufy V, Soltanalizadeh B, et al. A content-based literature recommendation system for datasets to improve data reusability - A case study on Gene Expression Omnibus (GEO) datasets. J Biomed Inform 2020;104:103399. [Crossref] [PubMed]
- Zail J, Eastwood K, Bugeja L, et al. Geo-mapping of young people in residential aged care. Australas J Ageing 2020;39:e288-94. [Crossref] [PubMed]
- Wu M, Shang X, Sun Y, et al. Integrated analysis of lymphocyte infiltration-associated lncRNA for ovarian cancer via TCGA, GTEx and GEO datasets. PeerJ 2020;8:e8961. [Crossref] [PubMed]
- Yu Y, Tian X. Analysis of genes associated with prognosis of lung adenocarcinoma based on GEO and TCGA databases. Medicine (Baltimore) 2020;99:e20183. [Crossref] [PubMed]
- Shen Y, Dong S, Liu J, et al. Identification of Potential Biomarkers for Thyroid Cancer Using Bioinformatics Strategy: A Study Based on GEO Datasets. Biomed Res Int 2020;2020:9710421. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods 2013;10:1209-10. [Crossref] [PubMed]
- Pathan M, Keerthikumar S, Ang CS, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597-2601. [Crossref] [PubMed]
- Dickinson A, Xu M, Silen S, et al. Newly detected DNA viruses in juvenile nasopharyngeal angiofibroma (JNA) and oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC). Eur Arch Otorhinolaryngol 2019;276:613-7. [Crossref] [PubMed]
- Bloebaum M, Poort L, Bockmann R, et al. Survival after curative surgical treatment for primary oral squamous cell carcinoma. J Craniomaxillofac Surg 2014;42:1572-6. [Crossref] [PubMed]
- Sandulache VC, Michikawa C, Kataria P, et al. High-Risk TP53 Mutations Are Associated with Extranodal Extension in Oral Cavity Squamous Cell Carcinoma. Clin Cancer Res 2018;24:1727-33. [Crossref] [PubMed]
- Kvedar JC, Manabe M, Phillips SB, et al. Characterization of sciellin, a precursor to the cornified envelope of human keratinocytes. Differentiation 1992;49:195-204. [Crossref] [PubMed]
- Champliaud MF, Burgeson RE, Jin W, et al. cDNA cloning and characterization of sciellin, a LIM domain protein of the keratinocyte cornified envelope. J Biol Chem 1998;273:31547-54. [Crossref] [PubMed]
- Chou CK, Fan CC, Lin PS, et al. Sciellin mediates mesenchymal-to-epithelial transition in colorectal cancer hepatic metastasis. Oncotarget 2016;7:25742-54. [Crossref] [PubMed]
- Kürklü E, Ozturk S, Cassidy AJ, et al. Clinical features and molecular genetic analysis in a Turkish family with oral white sponge nevus. Med Oral Patol Oral Cir Bucal 2018;23:e144-50. [Crossref] [PubMed]
- Takano M, Shimada K, Fujii T, et al. Keratin 19 as a key molecule in progression of human hepatocellular carcinomas through invasion and angiogenesis. BMC Cancer 2016;16:903. [Crossref] [PubMed]
- Song BJ, Abdelmegeed MA, Cho YE, et al. Contributing Roles of CYP2E1 and Other Cytochrome P450 Isoforms in Alcohol-Related Tissue Injury and Carcinogenesis. Adv Exp Med Biol 2019;1164:73-87. [Crossref] [PubMed]
- Wang Y, Millonig G, Nair J, et al. Ethanol-induced cytochrome P4502E1 causes carcinogenic etheno-DNA lesions in alcoholic liver disease. Hepatology 2009;50:453-61. [Crossref] [PubMed]
- Frank A, Seitz HK, Bartsch H, et al. Immunohistochemical detection of 1,N6-ethenodeoxyadenosine in nuclei of human liver affected by diseases predisposing to hepato-carcinogenesis. Carcinogenesis 2004;25:1027-31. [Crossref] [PubMed]
- Peccerella T, Arslic-Schmitt T, Mueller S, et al. Chronic Ethanol Consumption and Generation of Etheno-DNA Adducts in Cancer-Prone Tissues. Adv Exp Med Biol 2018;1032:81-92. [Crossref] [PubMed]
- Millonig G, Wang Y, Homann N, et al. Ethanol-mediated carcinogenesis in the human esophagus implicates CYP2E1 induction and the generation of carcinogenic DNA-lesions. Int J Cancer 2011;128:533-40. [Crossref] [PubMed]
- Abdel-Rahman O. Impact of baseline characteristics on extensive-stage SCLC patients treated with etoposide/carboplatin: A secondary analysis of a phase III study. Clin Respir J 2018;12:2519-24. [Crossref] [PubMed]
- Sattar A, Masood M, Nisar H, et al. Disease Characteristics and Treatment Outcome of Testicular Germ Cell Tumors Treated with Platinum-Based Regimens. J Coll Physicians Surg Pak 2018;28:292-6. [Crossref] [PubMed]
- Vijenthira A, Chan K, Cheung MC, et al. Cost-effectiveness of first-line treatment options for patients with advanced-stage Hodgkin lymphoma: a modelling study. Lancet Haematol 2020;7:e146-56. [Crossref] [PubMed]
- Patel P, Lavoratore SR, Flank J, et al. Chemotherapy-induced nausea and vomiting control in pediatric patients receiving ifosfamide plus etoposide: a prospective, observational study. Support Care Cancer 2020;28:933-8. [Crossref] [PubMed]
- Li H, Cimino SK. Clinical impact of the etoposide injection shortage. J Oncol Pharm Pract 2020;26:187-92. [Crossref] [PubMed]





