
Identification of a novel circRNA, hsa_circ_0065898, that regulates tumor growth in cervical squamous cell carcinoma
Introduction
Cervical squamous cell carcinoma (CSCC) is a common cancer; it is the fourth most frequently diagnosed cancer and fourth leading cause of cancer-related death in women (1). Although many novel treatment methods and technologies have been used in CSCC, the clinical outcomes of patients have not been satisfactory (2). Some studies have reported that the 5-year overall survival of CSCC patients is still less than 17% (3). In addition, specific and efficient treatments are lacking. Therefore, there is an urgent need to explore the mechanisms underlying CSCC.
MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) play an important role in tumor biology (4-6). Some reports have shown that circRNAs are involved in various human diseases, particularly cancers (7-10). CircRNAs act as “sponges” for miRNAs regulating gene expression at the post-transcriptional level. Zhang et al. showed that circular RNA circSATB2 promotes progression of non-small cell lung cancer cells (11). CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p (12).
Hsa_circ_0065898, located in the chromosomal region 3p21.2, is also known as circDCAF1. However, the molecular mechanism of hsa_circ_0065898 in CSCC remains unclear. In this study, we showed that circDCAF1 promoted CSCC proliferation and invasion, and constructed a regulatory network of circRNA targeting miRNAs-mRNAs. This study provides evidence for the treatment and pathogenesis of CSCC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2808).
Methods
The workflow was shown in Figure 1.
Downloading GSE102686 and identification of differentially expressed circRNAs
We searched the keywords “cervical cancer circRNA” in the National Centre of Biotechnology Information (NCBI) Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/), and downloaded the circRNA expression profiles in GSE102686, which included 5 CSCC tissues and 5 normal tissues. Differentially expressed circRNAs were identified using R and Perl software. P values <0.05 and log fold change (FC) >2 were selected to identify differentially expressed circRNAs. The standard names of circRNAs were converted using the Perl software. The standard names of circRNAs are shown in an ID txt file.
Target gene prediction and regulatory network construction
miRNAs targeting hsa_circ_0065898 were obtained from the circBase database (http://www.circbase.org/). The miRNAs are shown in an miRNA txt file. miRNA expression validation and association with prognosis of CSCC were downloaded from the online Kaplan-Meier (KM) Plotter (http://kmplot.com/analysis/). The genes targeted by miRNAs were predicted using miRDB.tsv, miRTarBase.tsv, and TargetScan.tsv. The regulatory network of circRNA-miRNAs-mRNAs and hub genes was constructed using Cytoscape software 3.6.0 (13,14). The CytoHubba application, a Cytoscape plugin, was used to select hub genes (15).
KM Plotter database analysis of miRNAs
The KM plotter database is capable in assessing the effect of genes and miRNAs on survival in 21 cancer types. The miRNA subsystems include 21 different cancer types. The primary purpose of the KM plotter is a meta-analysis-based discovery and validation of survival biomarkers (16). KM plotter was used to assess the prognostic value of microRNAs in CSCC. All cases were classified into a low expression group and a high expression group. KM survival plots, the hazard ratio (HR), 95% confidence interval (CI), and log rank P value were automatically shown on the webpage. A log rank P value <0.05 was considered statistically significant.
Cell culture
A CSCC cell line (SiHa) was purchased from the company Cosmo Bio (Tianjin, China). SiHa cells were cultured in F-12K and DMEM-H (Gibco, USA). All cells were cultured at 37 °C for 18–24 h in a humidified incubator containing 5% CO2. The expression level of hsa_circ_0065898 was tested by quantitative PCR (qPCR), as described in a previous study (7).
CCK-8-cell proliferation and transwell assays
To examine cell proliferation, migration and invasion ability, we conducted a transwell and CCK-8 assay as described previously (17).
qRT-PCR analysis
qRT-PCR was performed as previously described (18). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls. The primer sequences used were as follows: Si-NC: F: 5'-UUCUCCGAACGUGUCACGUTT-3'; R: 5'-ACGUGACACGUUCGGAGAATT-3'. Si-RNA: F: 5'-GAAGCUCUAUAAUGUGUUUAG-3'; R: 5'-AAACACAUUAUAGAGCUUCAG-3'. GAPDH: F: 5'-GGTGAAGGTCGGTGTGAACG-3'; R: 5'-CTCGCTCCTGGAAGATGGTG-3'. hsa_circ_0065898: F: 5'-CCGGGAAGCCAATGAAGATG-3'; R: 5'-CCAAAGTGCAGACAAAGGCT-3'.
Statistical analyses
The hsa_circ_0065898 expression and difference of data in different groups were investigated by t-test or one-way ANOVA followed by Tukey’s test, using GraphPad Prism 6.0 software (GraphPad Inc., La Jolla, CA, USA). Survival analysis was performed using the KM plotter database. P<0.05 was considered significant (*, P<0.05; **, P<0.01; ***, P<0.001). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Differentially expressed circRNA identification
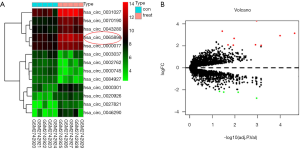
GSE102686 from the GEO database was analyzed, which included data from 5 CSCC tissues and five adjacent normal tissues. Among 13 differentially expressed circRNAs, 4 circRNAs were downregulated, while 9 circRNAs were upregulated. The four downregulated circRNAs were hsa_circ_0000745, hsa_circ_0084927, hsa_circ_0002762, and hsa_circ_0003037. The nine upregulated circRNAs were hsa_circ_0065898, hsa_circ_0070190, hsa_circ_0000077, hsa_circ_0031027, hsa_circ_0043280, hsa_circ_0027821, hsa_circ_0000301, hsa_circ_0020926, and hsa_circ_0046290. The results are presented in the volcano plot and heat map in Figure 2.
Regulatory network construction
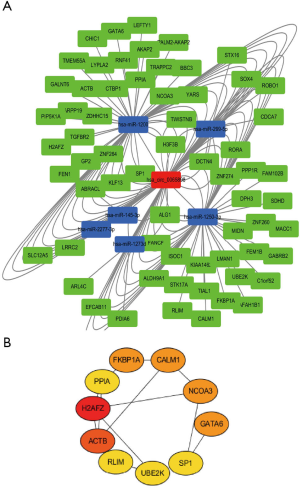
We used the keyword “hsa_circ_0065898” in the circBase database. We obtained the miRNAs that bind to the circRNA hsa_circ_0065898. The miRNAs binding hsa_circ_0065898 were hsa-mir-1200, hsa-mir-145, hsa-mir-1250, hsa-mir-1273d, hsa-mir-2277, and hsa-mir-299. In the present study, we used the type and network txt files to construct the circRNA-miRNA-mRNA regulatory network using the Cytoscape software 3.6.0. The relationship of hsa_circ_0065898 with miRNAs and genes is shown in Figure 3A. H2AFZ, ACTB, RLIM, UBE2K, SP1, GATA6, NCOA3, CALM1, FKBP1A, and PPIA were the hub genes (Figure 3B).
K-M Plotter database analysis
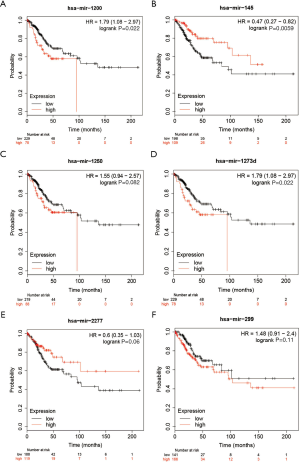
A total of 307 CSCC patients were included in the KM plotter database. The associations between the expression of hsa-mir-1200, hsa-mir-145, hsa-mir-1250, hsa-mir-1273d, hsa-mir-2277, and hsa-mir-299 and overall survival were analyzed. We found that the expression of hsa-mir-1200 (HR =1.79, 95% CI: 1.08–2.97, P=0.022), hsa-mir-1250 (HR =1.55, 95% CI: 0.94–2.57, P=0.082), hsa-mir-1273d (HR =1.79, 95% CI: 1.08–2.97, P=0.022), and hsa-mir-299 (HR =1.48, 95% CI: 0.91–2.4, P=0.11) were associated with poor prognosis. However, overexpression of hsa-mir-145 was associated with better overall survival (HR =0.47, 95% CI: 0.27–0.82, P=0.0059). The results are shown in Figure 4.
GO enrichment analysis and KEGG pathway analysis
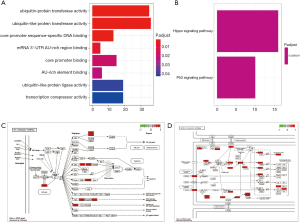
GO enrichment analysis and KEGG pathway analysis were performed on target genes using the R software and Perl software. GO functional enrichment analysis with a P value of 0.05 was obtained. The results are shown in Figure 5A. We found that “ubiquitin-protein transferase activity” was the most significant enrichment. The signaling pathways were significantly enriched in the Hippo signaling pathway and the p53 signaling pathway (Figure 5B,C,D).
hsa_circ_0065898 promotes proliferation, migration, and invasion
In the present study, we found that hsa_circ_0065898 was the most significantly expressed in GSE102686 samples from the GEO database (Figure 1). To further validate this result, we chose SiHa cell lines and found that the expression of hsa_circ_0065898 was significantly upregulated (Figure 6A).
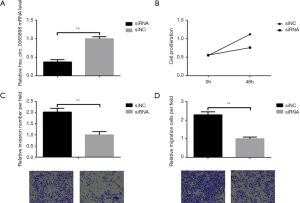
Upon evaluating the role of hsa_circ_0065898 on proliferation using the CCK-8 assay, we found that siRNA- hsa_circ_0065898 markedly inhibited cell proliferation (Figure 6B). The results of the transwell assay suggested that hsa_circ_0065898 significantly affected cell invasion and migration (Figure 6C,D).
Discussion
Cervical cancer is the second most prevalent cancer in women worldwide (19). CSCC accounts for a significant proportion of cervical cancer. The underlying mechanism of CSCC remains unclear. Thus, there is a need for novel diagnostic biomarkers for CSCC.
Non-coding RNAs, especially circRNAs, have been shown to regulate several types of cancer progression (20,21). Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway (18). Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression (22). In this study, hsa_circ_0065898 was highly expressed in CSCC cells. Knockdown of hsa_circ_0065898 markedly suppressed the proliferative rate. Our results showed that hsa_circ_0065898 promotes the progression of cervical cancer.
To further analyze the role of hsa_circ_0065898 in CSCC pathogenesis, we used the online circBase database to explore the direct interaction of hsa_circ_0065898 with miRNAs. Hsa-mir-1200, hsa-mir-145, hsa-mir-1250, hsa-mir-1273d, hsa-mir-2277, and hsa-mir-299 were the top 6 miRNAs selected to construct the regulatory network. hsa-mir-1200 showed low expression in osteosarcoma cells (23), and it played a role in arterial and venous endothelial cells exposed to gestational diabetes mellitus (24). hsa-mir-1200 showed a negative correlation with the grade of neuroendocrine tumor biology of the lung (25). This study found that hsa-mir-1200 was significantly associated with poor survival. hsa-mir-145 affected circular RNA expression in prostate cancer (26), glioblastoma cell (27), bladder cancer (28,29), and colorectal cancer (30). In CSCC, we showed that hsa-mir-145 overexpression had a better overall survival. hsa-mir-1250, hsa-mir-1273d, hsa-mir-2277, and hsa-mir-299 play roles in many diseases (31-33).
It has been reported that miRNAs can regulate tumor development by targeting mRNAs. In the present study, the hsa_circ_0065898-targeted miRNA–mRNA network may regulate ubiquitin-protein transferase activity, ubiquitin-like protein transferase activity, core promoter sequence-specific DNA binding, mRNA 3’-UTR AU-rich region binding, core promoter binding, AU-rich element binding, ubiquitin-like protein ligase activity, and transcription corepressor activity.
The p53 signaling pathway and the Hippo pathway affect tumor growth and progression (34,35). Zhang et al. found that inotodiol inhibited cell migration and invasion and induced apoptosis via a p53-dependent pathway in HeLa cells (36), and other study investigated the p53 signaling pathway regulated cervical cancer progression via miR-22/HDAC6 (37). He et al. showed that the Hippo/YAP pathway interacts with epidermal growth factor receptor (EGFR) signaling oncoproteins to regulate cervical cancer progression (38).
Kong et al. also found that the Hippo pathway played an important role in CSCC (39). In addition, we studied the main genes of the Hippo and p53 pathways, MST1, LATS1, LATS2, and P53, and found that they were associated with immune cell infiltration in CSCC.
Human Papillomavirus (HPV) plays an important role in cervical cancer. Epidemiological studies claim that human papillomavirus (HPV) infection is a necessary condition for cervical cancer development (40-42). However, the mechanism of HPV in cervical cancer need to be further studied.
In conclusion, the present study identified differentially expressed circRNAs and found that hsa_circ_0065898 could promote CSCC cell proliferation and invasion. We explored the regulatory network of hsa_circ_0065898 targeted miRNAs and genes, and our study provides more insights into the role of hsa_circ_0065898 in cervical cancer progression.
Conclusions
This study identified differentially expressed circRNAs and constructed the regulatory network of hsa_circ_0065898 targeting microRNAs and mRNAs. We demonstrated that hsa_circ_0065898 promoted CSCC cell proliferation, migration, and invasion. Hence, hsa_circ_0065898 might be useful as a biomarker for CSCC diagnosis and targeted therapy.
Acknowledgments
This manuscript has been released as a pre-print at Research Square. https://www.researchsquare.com/article/rs-29331/v1.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2808
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2808). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dai YF, Lin N, He DQ, et al. LZAP promotes the proliferation and invasiveness of cervical carcinoma cells by targeting AKT and EMT. J Cancer 2020;11:1625-33. [Crossref] [PubMed]
- Xia L, Yue Y, Li M, et al. CNN3 acts as a potential oncogene in cervical cancer by affecting RPLP1 mRNA expression. Sci Rep 2020;10:2427. [Crossref] [PubMed]
- Vedanayagam J, Chatila WK, Aksoy BA, et al. Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis. Nat Commun 2019;10:3682. [Crossref] [PubMed]
- Fattahi F, Kiani J, Khosravi M, et al. Enrichment of Up-regulated and Down-regulated Gene Clusters using Gene Ontology, miRNAs and lncRNAs in Colorectal Cancer. Comb Chem High Throughput Screen 2019;22:534-45. [Crossref] [PubMed]
- Zhou Y, Zheng X, Xu B, et al. The Identification and Analysis of mRNA-lncRNA-miRNA Cliques From the Integrative Network of Ovarian Cancer. Front Genet 2019;10:751. [Crossref] [PubMed]
- Wei J, Wang J, Gao X, et al. Identification of differentially expressed circRNAs and a novel hsa_circ_0000144 that promote tumor growth in gastric cancer. Cancer Cell Int 2019;19:268. [Crossref] [PubMed]
- Qiu L, Wang T, Ge Q, et al. Circular RNA Signature in Hepatocellular Carcinoma. J Cancer 2019;10:3361-72. [Crossref] [PubMed]
- Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy 2020;16:659-71. [Crossref] [PubMed]
- Chen J, Chen T, Zhu Y, et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res 2019;38:398. [Crossref] [PubMed]
- Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer 2020;19:101. [Crossref] [PubMed]
- Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer 2019;18:25. [Crossref] [PubMed]
- Wang J, Liu H, Xie G, et al. Identification of hub genes and key pathways of dietary advanced glycation end productsinduced nonalcoholic fatty liver disease by bioinformatics analysis and animal experiments. Mol Med Rep 2020;21:685-94. [PubMed]
- Zhang H, Zhong J, Tu Y, et al. Integrated Bioinformatics Analysis Identifies Hub Genes Associated with the Pathogenesis and Prognosis of Esophageal Squamous Cell Carcinoma. Biomed Res Int 2019;2019:2615921. [Crossref] [PubMed]
- Zhou Z, Li Y, Hao H, et al. Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis. Cell Transplant 2019;28:76S-86S. [Crossref] [PubMed]
- Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Zhang Z, Wang C, Zhang Y, et al. CircDUSP16 promotes the tumorigenesis and invasion of gastric cancer by sponging miR-145-5p. Gastric Cancer 2020;23:437-48. [Crossref] [PubMed]
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18:20. [Crossref] [PubMed]
- Guo J, Chen M, Ai G, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother 2019;115:108957. [Crossref] [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [Crossref] [PubMed]
- Riquelme I, Letelier P, Riffo-Campos AL, et al. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int J Mol Sci 2016;17:424. [Crossref] [PubMed]
- Huang X, Li Z, Zhang Q, Wang W, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer 2019;18:71. [Crossref] [PubMed]
- Li S, Pei Y, Wang W, et al. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle 2019;18:1281-91. [Crossref] [PubMed]
- Liu Y, Wang Y, Wang Y, et al. Gene expression changes in arterial and venous endothelial cells exposed to gestational diabetes mellitus. Gynecol Endocrinol 2020;36:791-5. [Crossref] [PubMed]
- Mairinger FD, Ting S, Werner R, et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: results of a profiling study. Mod Pathol 2014;27:1632-40. [Crossref] [PubMed]
- He JH, Han ZP, Zhou JB, et al. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J Cell Biochem 2018;119:9168-77. [Crossref] [PubMed]
- Kurogi R, Nakamizo A, Suzuki SO, et al. Inhibition of glioblastoma cell invasion by hsa-miR-145-5p and hsa-miR-31-5p co-overexpression in human mesenchymal stem cells. J Neurosurg 2018;130:44-55. [Crossref] [PubMed]
- Liu L, Wu SQ, Zhu X, et al. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma 2019;66:736-45. [Crossref] [PubMed]
- Wang J, Zhang C, Wu Y, et al. Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in bladder urothelial carcinoma. Cancer Cell Int 2019;19:327. [Crossref] [PubMed]
- Mao Z, Zhao H, Qin Y, et al. Post-Transcriptional Dysregulation of microRNA and Alternative Polyadenylation in Colorectal Cancer. Front Genet 2020;11:64. [Crossref] [PubMed]
- de Faria O Jr, Cui QL, et al. Regulation of miRNA 219 and miRNA Clusters 338 and 17-92 in Oligodendrocytes. Front Genet 2012;3:46. [Crossref] [PubMed]
- Kondybayeva capital A C, Akimniyazova A, et al. Prediction of miRNA interaction with mRNA of stroke candidate genes. Neurol Sci 2020;41:799-808.
- Duan H, Li X, Chen Y, et al. LncRNA RHPN1-AS1 promoted cell proliferation, invasion and migration in cervical cancer via the modulation of miR-299-3p/FGF2 axis. Life Sci 2019;239:116856. [Crossref] [PubMed]
- Hu L, Wang Y, Chen Z, et al. Hsp90 Inhibitor SNX-2112 Enhances TRAIL-Induced Apoptosis of Human Cervical Cancer Cells via the ROS-Mediated JNK-p53-Autophagy-DR5 Pathway. Oxid Med Cell Longev 2019;2019:9675450. [Crossref] [PubMed]
- Bi L, Ma F, Tian R, et al. AJUBA increases the cisplatin resistance through hippo pathway in cervical cancer. Gene 2018;644:148-54. [Crossref] [PubMed]
- Zhang SD, Yu L, Wang P, et al. Inotodiol inhibits cells migration and invasion and induces apoptosis via p53-dependent pathway in HeLa cells. Phytomedicine 2019;60:152957. [Crossref] [PubMed]
- Wongjampa W, Ekalaksananan T, Chopjitt P, et al. Suppression of miR-22, a tumor suppressor in cervical cancer, by human papillomavirus 16 E6 via a p53/miR-22/HDAC6 pathway. PLoS One 2018;13:e0206644. [Crossref] [PubMed]
- He C, Mao D, Hua G, et al. The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol Med 2015;7:1426-49. [Crossref] [PubMed]
- Kong F, Li Y, Hu E, et al. The Characteristic of S100A7 Induction by the Hippo-YAP Pathway in Cervical and Glossopharyngeal Squamous Cell Carcinoma. PLoS One 2016;11:e0167080. [Crossref] [PubMed]
- Shokar NK, Doan A, Calderon-Mora J, et al. The Prevalence of Genital Human Papillomavirus Subtypes in a Cohort of Hispanic Women Presenting for Cervical Cancer Screening Along the US-Mexico Border. Cancer Control 2020;27:1073274820951780. [Crossref] [PubMed]
- Palmer T, Cuschieri K. Human Papillomavirus Immunization and the Elimination of Cervical Carcinoma. Ann Intern Med 2020;173:935-6. [Crossref] [PubMed]
- Miranda PJC, Chagas BS, Coêlho M, et al. Correlation between human papillomavirus infection and histopathological diagnosis of women in Northeast Brazil. J Med Virol 2020; Epub ahead of print. [Crossref] [PubMed]



