
Transcriptome Sequencing reveals the expressed profiles of mRNA and ncRNAs and regulate network via ceRNA mediated molecular mechanism of lung adenocarcinoma bone metastasis in Xuanwei
Introduction
Lung cancer is one of the most malignant tumors that poses a threat to the health and life of the population (1). Its morbidity and mortality are the fastest growing, which is the main cause of the global cancer death rate (2). Among them, non-small cell lung cancer (NSCLC) account for 85% of lung cancer (3). For patients with early-stage NSCLC who have no evidence of mediastinal invasion or distant metastasis before surgery, surgical resection of the lung mass and the collection of mediastinal lymph nodes is the best treatment. However, the recurrence rate after surgery is very high (4,5). NSCLC can be divided into three subtypes of lung adenocarcinoma (LuAC), squamous cell carcinoma and large cell carcinoma. The most ordinary subtype of lung cancer is LuAC, which is characterized by strong metastatic ability and high mortality (6). The LuAC rates in Xuanwei, China, are among the highest around the world, especially in nonsmoking women (7). Various of risk factors for LuAC rates are reported, such as smoking, diet, atmospheric pollution, genetic factors, etc. Among them, smoky coal for cooking and heating is considered as a major risk factors in its pathogenesis (7). But because of all kinds of reason, the death rate hasn’t to be controlled effectively (8). And the patients are already at an advanced stage when they are diagnosed due to the scarcity of effective biomarkers for early detection (9). Several newer targeted therapies may shed new light on potential therapeutic breakthroughs due to surgical limitations, chemotherapy resistance, radiotherapy side effects and other factors, such as PD-1 noncoding RNAs (ncRNAs) and so on (10-13). Simultaneously, bone metastases are common in patients with (LuAC and are commonly osteolytic in nature (14). The survival time of lung cancer patients after bone metastasis is 6–10 months, while the 1-year survival rate is only 40–50% after the clinical treatment (15,16). The study of associated driven genes in lung cancer bone metastasis will provide new insight into the mechanism, potential therapeutic targets, promote the prognosis and survival in clinic (17,18). Although bone metastasis has important clinical significance in lung cancer, its pathological mechanism is still indistinct. Therefore, taking Xuanwei LuAC as the core research object, in-depth study of the pathogenesis of bone metastasis and systematic screening of early diagnosis and potential treatment targets will surely provide vital basis for clinical and scientific research.
Noncoding RNAs (ncRNAs) are central components of the human transcriptome that functionally regulate protein expression (19). The ncRNAs include long ncRNAs (lncRNAs, >200 bp, up to 100 kb), microRNAs (miRNAs <200 bp) and circular RNAs (circRNA, closed loop structure) (20,21). Competing endogenous RNAs (ceRNAs) hypothesis reveals a new mechanism for interaction between RNAs. MiRNAs can cause gene silencing by binding to mRNA. However, ceRNA can regulate gene expression by competitively binding to miRNAs. LncRNAs or circRNAs competitively binds to miRNA and acts as a sponge to regulate gene expression. CeRNA can bind miRNAs through miRNA response elements (MREs), affecting miRNA-induced gene silencing, thus leading to elevated levels of miRNA target genes (22,23). It has been reported that they play key roles in varieties of biological processes such as angiogenesis, cell proliferation, differentiation, apoptosis, autophagy, and immune response (24-28). The long non-coding RNA activated by TGF-β, lncRNA-ATB, acts as a ceRNA and competes with ZEB1 and ZEB2 to bind to miR-200 to drive the invasion and metastasis cascade of hepatocellular carcinoma (29). CircRNA_100290 can play a part in oral cancer by functioning as a sponge of miR-29 (30). ZEB1 initiates miR-181b-regulated ceRNA network to drive LuAC metastasis (31). LncRNA metastasis-associated LuAC transcript1MALAT1, which is a ceRNA that regulates autophagy-associated 7 (ATG7) gene expression via the sponge miR142-3p (32). As far as we all know, the ceRNA regulatory networks and its basic mechanism in LuAC bone metastasis have not been completely inquired.
High-through put transcriptome sequencing has been widely used to analyse particularly gene expression modes at different developmental phases. In present study, we investigated the mRNA, miRNA, lncRNA and circRNA expression in primary LuAC and LuAC bone metastasis in Xuanwei, China using RNA-sequencing. We structured the ceRNA network to detail the interactions or potential crosstalk among the key miRNA, mRNA, lncRNA and circRNA in primary LuAC and LuAC bone metastasis. Furthermore, through the integrated prediction and research of the ceRNA mechanism, we hope to screen a group of ncRNAs as biomarkers for the diagnosis of LuAC bone metastasis and come out a novel diagnostic and therapeutic strategy against LuAC bone metastasis in Xuanwei.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2376).
Methods
Study population
We used a retrospective cohort study design. In our study, the Xuanwei patients come from the Third Affiliated Hospital of Kunming Medical University. The detailed clinical data of patients are shown in Table 1. Fresh tumor tissue samples were procured from 3 cases of primary LuAC (P1-P3) and 3 cases of LuAC bone metastasis (M1-M3) after surgical resection. Tissue samples were frozen in liquid nitrogen prior to RNA extraction and stored at −80 °C. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Third Affiliated Hospital of Kunming Medical University (Yunnan Cancer Hospital) (No. KMUYXLL0046) and informed consent was taken from all the patients.
Table 1
| Index | P1 | P2 | P3 | M1 | M2 | M3 |
|---|---|---|---|---|---|---|
| Age, years | 50 | 73 | 64 | 43 | 74 | 65 |
| Gender | Male | Female | Female | Female | Male | Female |
| Site of metastasis | – | – | – | Bone | Bone | Bone |
RNA separation extraction and quality monitoring
RNA extraction of all tissue samples was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manual instruction. The RNA quality monitoring was performed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
MiRNA library construction, sequencing, and data processing
The library was constructed that used the Truseq RNA sample Prep Kit (Illumina, Inc., San Diego, CA, USA). The adapter is connected to the 3’ end of the RNA and then to the 5’ adapters. Subsequently, the RNA was fragmented into short base pairs when added the lysis buffer. Synthesis of single-stranded cDNA was performed by RNA reverse transcription. Amplification of miRNA high-throughput sequencing libraries was performed by PCR. Sequencing was performed by a lumina HiSeq4000 (Illumina, San Diego, CA, USA) platform.
The internal pipeline processes raw data (original reads), including adapter pruning, read alignment, and read counts. By using the popular alignment tool Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) to map the trimmed reads (also known as clean reads) to the human reference genome GRCh38 (33). Then, using the software miRDeep2 (https://www.mdcberlin.de/8551903/en/) to quantify the expression of miRNA (34). miRBase downloaded mature miRNA and miRNA precursors. Moreover, DEGseq package in R was used to identify DEmiRNAs between samples. The cut-off criteria are P value <0.05 and |log2 (Fold_change)| >2.
MiRNA and lncRNA sequencing and data processing
The library construction standards were as follows: (I) concentration of RNA ≥200 ng/mL; (II) RNA with amount >5 µg; (III) RIN >7; (IV) 1.8< OD260/280 <2.2. Fragmentation of rRNA-deleted RNA after removal of ribosomal RNA, using the Truseq RNA sample Prep Kit (Illumina, Inc., San Diego, CA, USA) was got to structure the cDNA library. Sequencing of libraries were performed by an Illumina HiSeq x-ten (Illumina Inc., San Diego, CA, USA) platform.
All the raw data obtained from high-throughput RNA-sequencing was translated into raw FASTQ sequence data by using Base Calling. To acquire the clean reads from the RNA sequencing results of lncRNA and mRNA, we used cutadapt (http://cufflinks.cbcb.umd.edu/) and Fastx-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to remove lower quality sequences, including adapter sequences, sequences with mass fraction <20 and sequences with N base rates of raw reads >10% and sequence less than 18 bp. The mapped reads were quantified with cuffquant, via using the Cufflinks (http://cufflinks.cbcb.umd.edu/) package of Cuffdiff program identify the DEmRNAs and DElncRNAs between samples. The P value <0.05, with |log2 (Fold-change)| >2 as the cutoff criteria.
CircRNA library construction, sequencing, and data processing
Removing the ribosomal RNA from total RNA, Digestion of linear RNA with RNaseR was carried out to isolate circRNAs. Then, the cDNA was constructed by further adding the reverse transcriptase to the RNA sample with RNase R. The 3’end of the cDNA is ligated with the base “A” and ligated to the linker, then the fragment size is selected, and finally PCR amplification is performed. Sequencing of libraries were performed by an IlluminaHiSeq x-ten (Illumina Inc., San Diego, CA, USA) platform.
Raw reads of FASTQ format were then processed through in-house Perl scripts and removed low quality reads using software SOAPnuke. Align clean RNA sequencing reads with reference genome, using CIRI (v2.0.5) and find circ (v1.2) predict circRNA. After combining the results of the two softwares, quantitative and differential expression analysis of circRNA was performed. We analyzed the circRNA abundance by using the normalized reads count. DEcircRNAs were evaluated by DESeq2 (R software package) (2). In the differential expression analysis, P value <0.05, with |log2 (Fold_-change)| >2 was as the cutoff criteria.
Functional analysis of DEmRNAs and DEncRNAs
To explore the functions of DEmRNAs and DEncRNAs, using the annotation of gene ontology (GO) and analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways. However, for the differential expression genes, we use GO analysis (http://www.geneontology.org) to create hierarchical categories based on molecular function (MF), cellular composition (CC), and biological process (BP) characteristics to elucidate genes. In addition, KEGG (http://www.genome.jp/kegg/) was used to analyse the important pathway in DEmRNAs and DEncRNAs. Using Metascape (http://metascape.org/gp/index.html) bioinformatics resources to perform GO functional annotation and KEGG pathway analysis of differentially expressed genes. P<0.001 was selected as the cutoff standards of significance.
Analysis of the ceRNA regulatory network
The ceRNA analysis was performed on mRNAs, lncRNAs and circRNAs with significant correlations in expression levels, searching for potential MREs in the lncRNA, circRNA and mRNA sequences. The lncRNA/circRNA-miRNA-mRNA interaction was predicted by considering the overlap of lncRNA/circRNA with the same miRNA seed sequence binding site in the mRNA sequence. The miRNA binding sites were predicted by miRcode (http://www.mircode.org/) and TargetScan (http://www.targetscan.org/) predicts the miRNA-mRNA interactions.
QRT-PCR verify differential gene expression
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the instructions. Reverse transcription into cDNA using reverse Transcriptase Kit (TaKaRa, Dalian, China) using mRNA as a template. The miRNA was reverse transcribed using the miRcute miRNA First-strand cDNA Synthesis Kit (TaKaRa, Dalian, China). Designed the forward and reverse primers, qRT-PCR detection was performed on a BIO-RAD IQ5 RT-PCR detection system (Bio-Rad Laboratories Inc., Germany). Primer sequences of qRT-PCR reactions were shown in Table S1. Calculating the relative expression of DEmRNAs and DEncRNAs used the 2−ΔΔCt method. GAPDH was used as control. The lncRNAs, circRNAs and miRNAs expression levels were normalized against the snU6.
Statistical analysis
All statistical analyses were performed by GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA). Two independent sample t test was used to compare the mean of measurement data between groups. And the data was represented as means with standard deviation (SD). α=0.05 was used as the statistical test level in this study.
Results
Identification of DEmRNAs and DEncRNAs
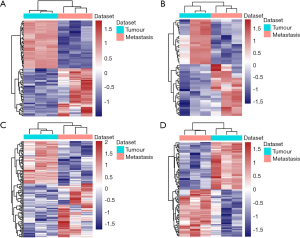
Significant analysis of RNA sequencing was performed using Cuffdiff software and analyse differential expressed mRNAs and ncRNAs the standards of P<0.05. The clustering heatmap of DEmRNAs, DEmiRNAs, DElncRNAs and DEcircRNAs were showed in Figure 1 separately. In all of 2,141 DEmRNAs (1046 up-regulated and 1095 down-regulated DEmRNAs) were confirmed in the Xuanwei patients with primary LuAC vs. LuAC bone metastasis, respectively (Figure 1A). In addition, there are 43 DEmiRNAs, including 23 upregulated and 20 downregulated miRNAs and 136 DElncRNAs, including 76 upregulated and 60 downregulated lncRNAs; 706 DEcircRNAs, including 365 upregulated and 351 downregulated circRNAs in the Xuanwei patients with primary LuAC vs. LuAC bone metastasis, respectively (Figure 1B,C,D).
Functional annotation of DEmRNAs and DEncRNAs
To investigate the function of ncRNAs in the Xuanwei patients with LuAC bone metastasis, we used GO annotation and KEGG pathway analysis to identify the functions of DEmRNAs and DEncRNAs between LuAC bone metastasis and primary LuAC patients in Xuanwei. LncRNAs might regulate the nearby protein-coding genes expression. In the three major GO categories of biological process, molecular function and cellular component, there are overly representative terms in GO analysis. The top 15 outcomes of gene-annotated DEmRNAs and DEncRNAs were analyzed for biological processes (BP), cellular components (CC) and molecular function (MF). Sort by P value and select the prediction term with P value <0.05. Based on the prediction of the dissemination of the target gene in GO, the gene functions of DEncRNAs were clarified. Each GO item enrichment statistically analyzes the number of genes and is represented in the form of histograms.
The most enriched BPs were cell division and cell adhesion that were showed by the GO analysis of DEmRNAs. The proteinaceous extracellular matrix, plasma membrane, chromosome and spindle microtubule were the meaningfully enriched in the CCs. The single-stranded DNA-dependent ATPase activity, microtubule motor activity and extracellular matrix structural constituent were the importantly enriched in the MF (Figure 2A).
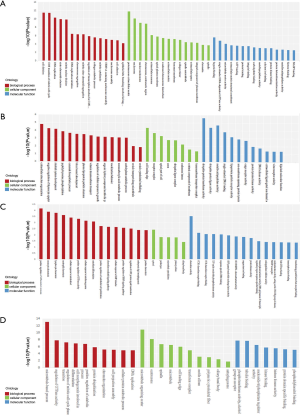
GO analysis for the targeted gene of DEmiRNAs revealed that the most expressively enriched BPs were extracellular structure organization, negative regulation of thymocyte apoptotic process, skeletal system development and ceramide biosynthetic process. The cell leading edge, receptor complex, apical part of cell and axon were the most noteworthy enriched in the CCs. The ubiquitin-protein transferase activity, metalloendopeptidase activity, single-stranded DNA binding and lipoprotein particle receptor activity were the most meaningfully enriched in the MF (Figure 2B).
The GO analysis for nearby protein-coding genes of DElncRNAs revealed that the most significantly enriched BPs were positive regulation of inflammatory response, positive regulation of GTPase activity and osteoblast differentiation. The microtubule cytoskeleton, primary cilium and ciliary basal body were the most significantly enriched in the CCs. The chemokine activity3’,5’-cyclic-nucleotide phosphodiesterase activity and transforming growth factor beta receptor binding were the importantly enriched in the MF (Figure 2C).
The GO analysis for the source gene of DEcircRNAs revealed that the most noteworthy enriched BPs were microtubule-based process, regulation of GTPase activity, regulation of mitotic cell cycle phase transition and cell morphogenesis involved in differentiation revealed that the most expressively enriched in the BPs. The microtubule organizing center, centrosome, spindle, microtubule, cell leading edge and transferase complex were the importantly enriched in the CCs. The phosphotransferase activity, alcohol group as acceptor, tubulin binding, nucleoside-triphosphatase regulator activity and kinase binding were the most noticeable enriched in the MF (Figure 2D).
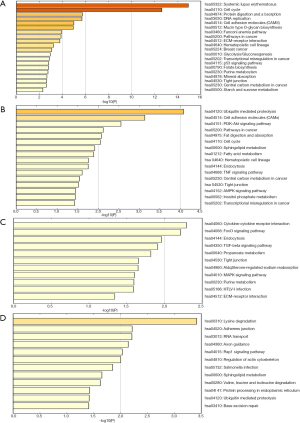
To acquire further comprehensions of the different biological functions of DEmRNAs and DEncRNAs, we performed KEGG pathway analysis. The top 20 pathways in the DEmRNAs are shown (Figure 3A, Table S2). Intriguingly, some of identified KEGG pathways comprised cell adhesion molecules (CAMs), pathways in cancer, ECM-receptor interaction and p53 signaling pathway. The total 16 pathways in the target mRNAs were based on the DEmiRNAs were shown (Figure 3B, Table S3). The most meaningfully associated pathways were PI3K-Akt signaling pathway, cell cycle, TNF signaling pathway and AMPK signaling pathway. The total 11 pathways in the nearby mRNAs of DE lncRNAs were shown (Figure 3C, Table S4). The most notably pathways were FoxO signaling pathway, endocytosis, TGF-beta signaling pathway and MAPK signaling pathway. The total 12 pathways in the source gene of DEcircRNAs were shown (Figure 3D, Table S5). The importantly pathways were adherens junction, Rap1 signaling pathway and regulation of actin cytoskeleton. These pathways are suggested to be involved in the metastatic mechanisms of LuAC bone metastasis.
Construction of ceRNA regulatory network
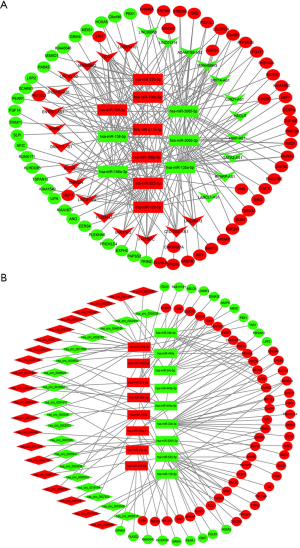
The study found that RNA regulates MREs through a mechanism called the ceRNA hypothesis. MREs involved in ceRNA networks can regulate mRNA expression and have also been discovered (35). The role of lncRNAs and circRNAs as miRNA sponges can inhibit the regulation of target genes on miRNAs, thereby indirectly regulating gene expression and functioning. Accordingly, lncRNA/circRNA-miRNA-gene and lncRNA/circRNA (as a sponge), miRNA (as the center) and mRNA (as the target) relationship pairs were established. From the high-throughput sequencing data of tissue samples, we integrated the expression details and regulatory relationships of the mRNAs, miRNAs, lncRNAs and circRNAs to construct a ceRNA network (Figure 4). Our results showed that the 67 mRNAs, 27 lncRNAs and 12 miRNAs were involved in the lncRNA-miRNA-mRNA ceRNA network (Figure 4A). However, the ceRNA networks were found on circRNA-miRNA-mRNA interactions include 40 circRNAs, 17 miRNAs and 71 mRNAs (Figure 4B). These ceRNA regulatory networks contain circRNAs/ lncRNAs, miRNAs and mRNAs, which may play a key role in the LuAC in Xuanwei with bone metastasis.
QRT-PCR confirmed the ncRNAs and mRNA expression
Through bioinformatics analysis and literature review, we finally selected 12 significantly different candidate genes to verify by qRT-PCR in Xuanwei LuAC samples. We verified the expression of 2 DEcircRNAs (circ_0000053 and circ_00001495), 2 DElncRNAs (ADAMTS9-AS2 and TEX41), 4 DEmiRNAs (miR-196b-5p, miR-223-3p, miR-135a-5p and miR-9) and 4 DEmRNAs (MEIS1, HOXA5, SCARB1 and RUNX2) (Figure 5). Compared with Xuanwei patients with LuAC and normal, the expression of circ_0000053, circ_00001495, TEX41, miR-223-3p, miR-9, RUNX2, SCARB1 and ADAMTS9-AS2, miR-135a-5p, MEIS1, HOXA5 were detected significantly upregulated and down regulated in Xuanwei patients with LuAC bone metastasis (Figure 5). Meanwhile, in this study, the expression of these eleven genes was united with the RNA-seq results (Table S6).
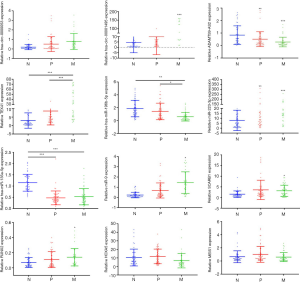
However, in our study, the expression of miR-196b-5p was inconsistent with that in RNA-seq results (Figure 5, Table S6).
Discussion
Recent study shows that, Xuanwei has the highest morbidity and mortality of LuAC in China. And the rates are also rather high among non-smoking women (36). Bone is a common transfer site in cancer patients with LuAC. Around 30–40% of patients with advanced lung adenocarcinoma cancer occur bone metastases during their onset, this will lead a remarkable negative influence on morbidity and survival (37). The presence of bone metastases suggests a poor prognosis, with a 1-year survival rate of 40% to 50% and a median survival time of only 6 to 10 months (38,39). In recent years, non-coding RNAs have been found in multiple cancers and are connected with comprehensive biological regulatory functions (40). Circulating non-coding RNAs in blood (serum, plasma) as important potential of biomarkers in liquid biopsies for diagnosis and follow-up of patients with all kinds of tumors (41,42).This study used a next-generation sequencing technique to quantify and comprehensively analyze the coding and non-coding transcriptomes of primary LuAC and LuAC bone metastasis in Xuanwei. Our analyses proved remarkable differences in the expression patterns of mRNA, lncRNA, circRNA and miRNA in primary LuAC and LuAC bone metastasis in Xuanwei, and dynamic changes in DEmRNA, DElncRNA, DEmiRNA and DEcircRNAs. We first found that the expression patterns of mRNAs and ncRNAs and are more appropriate for the identification of primary LuAC and LuAC bone metastasis samples in Xuanwei.
Overall, our result has proved that mRNAs, miRNAs, circRNAs and lncRNAs different populations participated in the pathogenesis of LuAC bone metastasis in Xuanwei. Unsupervised hierarchical clustering analysis the expression profiles of DElncRNAs, DEcircRNAs, DEmiRNAs and DEmRNAs, it was found that mRNA and ncRNAs expression profiles can largely distinguish between the primary LuAC and LuAC bone metastasis in Xuanwei. Using GO and KEGG pathway analyze the potential functions of DEmRNAs and DEncRNAs in the Xuanwei LuAC bone metastasis. In addition, according to these findings, we suggested that ceRNAs play an important role in Xuanwei LuAC bone metastasis and the ceRNA regulatory networks were established. We found that most of the DEmiRNAs may be associated with Xuanwei LuAC metastasis mechanism. However, due to the lack of research, majority of the DElncRNAs/DEcircRNAs were with unknown function.
A total of 2,141 DEmRNAs, 43 DEmiRNAs, 136 DElncRNAs and 706 DEcircRNAs were identified in both primary LuAC and LuAC bone metastasis in Xuanwei. DEmRNAs and DEncRNAs were significantly enriched in ECM-receptor interaction, PI3K/Akt signaling pathway and TGF-β signaling pathway (43). Previous studies also indicated that the dysregulation of ncRNAs and mRNA may contribute to Xuanwei LuAC metastasis. MiR-196b as a tumor suppressor target down-regulate Runx2 and regulate the PI3K/AKT/GSK3β, JNK and Smad signal pathways to suppress lung cancer cells growth and metastasis (44). MiR-130a-5p directly targeted RUNX2 regulates tumor invasive and metastatic potential in NSCLC (45). MiR-449a as a tumor suppressor in NSCLC and its function is through targeting HMGB1-mediated NF-κB signaling pathway, induced suppression of growth, migration and invasion (46). MiR-370 may bind to the 3’UTR of EGFR to restrain EGFR expression, inhibition of growth, angiogenesis and metastasis of NSCLC by down-regulating ERK1/2 and AKT signaling pathways (47). MiR-196a as a key mediator, through regulating FoxO1, CDKN1B and HOXA9, mediated the proliferative, pro-migratory and tumorigenic activity of lung cancer cells (48). MiR-223-3p directly inhibited the expression ofNLRP3 and IкB kinaseα activity and these two important mediators are thought to be associated with I/R-induced cell necroptosis and inflammation in the treatment of ischemic heart disease (49). MiR-223-3p regulated RIPK3 expression by directly targeting the 3’UTR of RIPK3, up-regulated 3-MCPD-dipalmitate-induced AKI and activated RIPK1/RIPK3/MLKL associated necrotic pathway in acute kidney injury (50). MiR-223-3p targets FOXO3 and promotes osteogenic differentiation of bone marrow mesenchymal stem cells by enhancing autophagy (42). MiR-9 regulates ferroptosis in melanoma cells by targeting GOT1, indicating the crucial role of miRNA in ferroptosis (51). In our current study, we showed that miR-196a-5p, miR-9-3p and miR-223-3p were upregulated in Xuanwei LuAC bone metastasis. Therefore, these miRNAs may act as important roles in Xuanwei LuAC metastasis mechanism, ferroptosis and necroptosis. MiR-196a can enhances cell migration, invasion and EMT by regulating the expression of HOXA5 in osteosarcoma (52). However, the expression of miR-196b-5p was inconsistent with that in RNA-seq results in our study (45). The reason may be related to the expansion of the sample size in the qRT-PCR and the selection of serum as the test sample. However, the function of miR-196b-5p in Xuanwei LuAC bone metastasis and its mechanism need further experimental verification.
Most of the DElncRNAs/DEcircRNAs were few studied. High expression of lncRNA ADAMTS9-AS2 in lung cancer cells obviously could reduce proliferation ability and inhibit migration, as well as promoting their apoptosis rate (43). The lncRNATEX41 was the variants of AS and affect the pathogenesis of aortic valve stenosis (53). TEX41-miR-340-COMMD6 ceRNA network to play an important role in head and neck squamous cell carcinoma (54). The non-coding RNA Linc00152 is a newly discovered lncRNA that has been reported to participate in tumor initiation and progression (55,56). LncRNA DLEU2 influences the proliferation, migration, and invasion of laryngeal cancer (57). In the breast cancer, hsa_circ_0002113 expression upregulated regulated breast cancer tumorigenesis and progression (58). However, lncRNA and circRNA of LuAC bone metastasis have little been reported until now.
Pseudogenes, circRNAs, lncRNAs and mRNAs can be used as ceRNAs because of RNA containing miRNA response elements. As natural miRNA sponges, they inhibit miRNA function by sharing miRNA response elements and coordinate with each other (59). In the process of LuAC bone metastasis, the underlying mechanism by which ncRNAs interact with mRNAs is unclear. Therefore, lncRNA/circRNA-miRNA-mRNA of LuAC bone metastasis was constructed that basing on the data analysis from next generation sequencing. These breakthrough discoveries will increase our understanding of the function of ncRNAs in the metastasis mechanism of LuAC bone metastasis. CeRNA network prediction and bioinformatics analysis are able to fully understand the ncRNA function, which may involve in the initiation and progression of the disease (60).
In our ceRNA network, we also found lncRNA ADAMTS9-AS2, TEX41, DLEU2 and LINC00152 were ceRNAs of miR-223-3p targeting SCARB1 (alias SR-B1). The SR-B1is a high-affinity HDL receptor that has been shown to mediate HDL-dependent lipid transport signaling in a variety of different cell types (61). The SCARB1 targeting of HPPS (HDL-mimicking peptide-phospholipid nanoscaffold (HPPS) nanoparticle composed of the cholesteryl oleate) plays an important role inefficient delivery in lysosomes (62). The research has identified a role of SR-B1 in regulating the protective effects of HDL against necrosis in myocardial necrosis (63) The circRNA-associated-ceRNA networks involved in the LuAC bone metastasis was selected. Hsa_circ_0000053 was ceRNAs of miR-196a-5p/miR-196b-5p targeting HOXA5. HOXA5 belongs to the Hox gene family encoding transcription factors known for its role in the formation of skeletal patterns (64). The dimer between HOX and PBX inhibits the necrotic pathway, possibly through inhibition of RIP kinase. HOX/PBX dimer also inhibit the expression of p21, which might in turn block apoptosis in acute myeloid leukemia cells (65). In summary, we speculated that lncRNA, circRNA and miRNA molecules may be pivotal regulators of varying Xuanwei LuAC course, such as metastasis and necroptosis.
Conclusions
In conclusion, our current research confirmed the extensive expression profile of ncRNAs in Xuanwei LuAC bone metastasis and provided a breakthrough data incorporation analysis of ceRNAs in Xuanwei LuAC with bone metastasis. A catalog of predicted ncRNAs in Xuanwei LuAC with bone metastasis was also defined. These rich data suggested that ceRNAs may regulate the expression of genes encoding related proteins and play a crucial role in Xuanwei LuAC with bone metastasis. Taken together, the findings further extended our knowledge of ceRNA biology and contributed to the understanding of their regulation roles in Xuanwei LuAC with metastasis mechanism and necroptosis. Regardless, our results offer worthy information for ncRNAs studies and clinical diagnosis, treatment, and prevention of Xuanwei LuAC.
Acknowledgments
Funding: This work was financially funded by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2376
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2376
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2376). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Third Affiliated Hospital of Kunming Medical University (Yunnan Cancer Hospital) (No. KMUYXLL0046) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fidler MM, Bray F, Soerjomataram I. The global cancer burden and human development: A review. Scand J Public Health 2018;46:27-36. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Zell JA, Ignatius Ou SH, Ziogas A, et al. Validation of the proposed International Association for the Study of Lung Cancer non-small cell lung cancer staging system revisions for advanced bronchioloalveolar carcinoma using data from the California Cancer Registry. J Thorac Oncol 2007;2:1078-85. [Crossref] [PubMed]
- Hoffman PC, Mauer AM, Vokes EEJL. Lung cancer. Lancet 2000;355:479-85. [Crossref] [PubMed]
- Vermeulen R, Downward GS, Zhang J, et al. Constituents of Household Air Pollution and Risk of Lung Cancer among Never-Smoking Women in Xuanwei and Fuyuan, China. Environ Health Perspect 2019;127:97001. [Crossref] [PubMed]
- Li R, Liu Y, Wang T, et al. The characteristics of lung cancer in Xuanwei County: A review of differentially expressed genes and noncoding RNAs on cell proliferation and migration. Biomed Pharmacother 2019;119:109312. [Crossref] [PubMed]
- Yang Y, Chen K, Zhou Y, et al. Application of serum microRNA-9-5p, 21-5p, and 223-3p combined with tumor markers in the diagnosis of non-small-cell lung cancer in Yunnan in southwestern China. Onco Targets Ther 2018;11:587-97. [Crossref] [PubMed]
- Zhang Y, Lin Q, Xu T, et al. Out of the darkness and into the light: New strategies for improving treatments for locally advanced non-small cell lung cancer. Cancer Lett 2018;421:59-62. [Crossref] [PubMed]
- Arbour KC, Luu AT, Luo J, et al. Deep learning to estimate RECIST in patients with NSCLC treated with PD-1 blockade. Cancer Discov 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Shih JH, Chen HY, Lin SC, et al. Integrative analyses of noncoding RNAs reveal the potential mechanisms augmenting tumor malignancy in lung adenocarcinoma. Nucleic Acids Res 2020;48:1175-91. [Crossref] [PubMed]
- Fang Y, Zhang C, Wu T, et al. Transcriptome Sequencing Reveals Key Pathways and Genes Associated with Cisplatin Resistance in Lung Adenocarcinoma A549 Cells. PLoS One 2017;12:e0170609. [Crossref] [PubMed]
- Ulas A, Bilici A, Durnali A, et al. Risk factors for skeletal-related events (SREs) and factors affecting SRE-free survival for nonsmall cell lung cancer patients with bone metastases. Tumour Biol 2016;37:1131-40. [Crossref] [PubMed]
- Tsuya A, Kurata T, Tamura K, et al. Skeletal metastases in non-small cell lung cancer: a retrospective study. Lung Cancer 2007;57:229-32. [Crossref] [PubMed]
- Kong P, Yan J, Liu D, et al. Skeletal-related events and overall survival of patients with bone metastasis from nonsmall cell lung cancer – A retrospective analysis. Medicine 2017;96:e9327. [Crossref] [PubMed]
- Rosen LS, Gordon D, Tchekmedyian NS, et al. Long-term efficacy and safety of zoledronic acid in the treatment of skeletal metastases in patients with nonsmall cell lung carcinoma and other solid tumors: A randomized, Phase III, double-blind, placebo-controlled trial. Cancer 2004;100:2613-21. [Crossref] [PubMed]
- Xu S, Yang F, Liu R, et al. Serum microRNA-139-5p is downregulated in lung cancer patients with lytic bone metastasis. Oncol Rep 2018;39:2376-84. [Crossref] [PubMed]
- Lanzafame M, Bianco G, Terracciano LM, et al. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int J Mol Sci 2018;19:682. [Crossref] [PubMed]
- Klinge, Carolyn MJERC. Noncoding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer 2018;25:R259-82. [Crossref] [PubMed]
- Xie L, Yao Z, Zhang Y, et al. Deep RNA sequencing reveals the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma tumorigenesis and pulmonary metastasis. Cell Death Dis 2018;9:772. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Chen C, Tan H, Bi J, et al. Identification of Competing Endogenous RNA Regulatory Networks in Vitamin A Deficiency-Induced Congenital Scoliosis by Transcriptome Sequencing Analysis. Cell Physiol Biochem 2018;48:2134-46. [Crossref] [PubMed]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 2012;441:523-40. [Crossref] [PubMed]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2007;96:R40-4. [PubMed]
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8. [Crossref] [PubMed]
- Mattick JS. Long noncoding RNAs in cell and developmental biology. Semin Cell Dev Biol 2011;22:327. [Crossref] [PubMed]
- Yang Z, Xie L, Han L, et al. Circular RNAs: Regulators of Cancer-Related Signaling Pathways and Potential Diagnostic Biomarkers for Human Cancers. Theranostics 2017;7:3106-17. [Crossref] [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene 2017;36:4551-61. [Crossref] [PubMed]
- Tan X, Banerjee P, Liu X, et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest 2018;128:1267-82. [Crossref] [PubMed]
- Song TF, Huang LW, Yuan Y, et al. LncRNA MALAT1 regulates smooth muscle cell phenotype switch via activation of autophagy. Oncotarget 2018;9:4411-26. [Crossref] [PubMed]
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [Crossref] [PubMed]
- Friedlander MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012;40:37-52. [Crossref] [PubMed]
- Guo LL, Song CH, Wang P, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol 2015;21:11680-7. [Crossref] [PubMed]
- Wong JYY, Downward GS, Hu W, et al. Lung cancer risk by geologic coal deposits: A case-control study of female never-smokers from Xuanwei and Fuyuan, China. Int J Cancer 2019;144:2918-27. [Crossref] [PubMed]
- Bae HM, Lee SH, Kim TM, et al. Prognostic factors for non-small cell lung cancer with bone metastasis at the time of diagnosis. Lung Cancer 2012;77:572-7. [Crossref] [PubMed]
- Al Husaini H, Wheatley-Price P, Clemons M, et al. Prevention and management of bone metastases in lung cancer: a review. J Thorac Oncol 2009;4:251-9. [Crossref] [PubMed]
- Niu YJ, Wen YT, Shen WW, et al. Risk factors for bone metastasis in patients with primary lung cancer: study protocol for a systematic review. BMJ Open 2014;4:e005202. [Crossref] [PubMed]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012;40:6391-400. [Crossref] [PubMed]
- Zeuschner P, Linxweiler J, Junker K. Non-coding RNAs as biomarkers in liquid biopsies with a special emphasis on extracellular vesicles in urological malignancies. Expert Rev Mol Diagn 2020;20:151-67. [Crossref] [PubMed]
- Long C, Cen S, Zhong Z, et al. FOXO3 is targeted by miR-223-3p and promotes osteogenic differentiation of bone marrow mesenchymal stem cells by enhancing autophagy. Hum Cell 2021;34:14-27. [Crossref] [PubMed]
- Liu C, Yang Z, Deng Z, et al. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life 2018;70:536-46. [Crossref] [PubMed]
- Bai X, Meng L, Sun H, et al. MicroRNA-196b Inhibits Cell Growth and Metastasis of Lung Cancer Cells by Targeting Runx2. Cell Physiol Biochem 2017;43:757-67. [Crossref] [PubMed]
- Ma F, Xie Y, Lei Y, et al. The microRNA-130a-5p/RUNX2/STK32A network modulates tumor invasive and metastatic potential in non-small cell lung cancer. BMC Cancer 2020;20:580. [Crossref] [PubMed]
- Wu D, Liu J, Chen J, et al. miR-449a Suppresses Tumor Growth, Migration, and Invasion in Non-Small Cell Lung Cancer by Targeting a HMGB1-Mediated NF-κB Signaling Pathway. Oncol Res 2019;27:227-35. [Crossref] [PubMed]
- Liu X, Huang YG, Jin CG, et al. MicroRNA-370 inhibits the growth and metastasis of lung cancer by down-regulating epidermal growth factor receptor expression. Oncotarget 2017;8:88139-51. [Crossref] [PubMed]
- Guerriero I, D'Angelo D, Pallante P, et al. Analysis of miRNA profiles identified miR-196a as a crucial mediator of aberrant PI3K/AKT signaling in lung cancer cells. Oncotarget 2017;8:19172-91. [Crossref] [PubMed]
- Qin D, Wang X, Li Y, et al. MicroRNA-223-5p and -3p Cooperatively Suppress Necroptosis in Ischemic/Reperfused Hearts. J Biol Chem 2016;291:20247-59. [Crossref] [PubMed]
- Huang G, Xue J, Sun X, et al. Necroptosis in 3-chloro-1, 2-propanediol (3-MCPD)-dipalmitate-induced acute kidney injury in vivo and its repression by miR-223-3p. Toxicology 2018;406-407:33-43. [Crossref] [PubMed]
- Zhang K, Wu L, Zhang P, et al. miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol Carcinog 2018;57:1566-76. [Crossref] [PubMed]
- Wang X, Zhang L, Zhao N, et al. MiR-196a promoted cell migration, invasion and the epithelial-mesenchymal transition by targeting HOXA5 in osteosarcoma. Cancer Biomark 2020;29:291-8. [Crossref] [PubMed]
- Helgadottir A, Thorleifsson G, Gretarsdottir S, et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9:987. [Crossref] [PubMed]
- Yang M, Huang W, Sun Y, et al. Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis. British Journal of Cancer 2019;121:699-709. [Crossref] [PubMed]
- Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol 2014;35:5441-7. [Crossref] [PubMed]
- Zhang PP, Wang YQ, Weng WW, et al. Linc00152 promotes Cancer Cell Proliferation and Invasion and Predicts Poor Prognosis in Lung adenocarcinoma. J Cancer 2017;8:2042-50. [Crossref] [PubMed]
- Xie ZZ, Xiao ZC, Song YX, et al. Long non-coding RNA Dleu2 affects proliferation, migration and invasion ability of laryngeal carcinoma cells through triggering miR-16-1 pathway. Eur Rev Med Pharmacol Sci 2018;22:1963-70. [PubMed]
- Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017;9:1175-88. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Liu W, Zhang J, Zou C, et al. Microarray Expression Profile and Functional Analysis of Circular RNAs in Osteosarcoma. Cell Physiol Biochem 2017;43:969-85. [Crossref] [PubMed]
- Gao M, Zhao D, Schouteden S, et al. Regulation of high-density lipoprotein on hematopoietic stem/progenitor cells in atherosclerosis requires scavenger receptor type BI expression. Arterioscler Thromb Vasc Biol 2014;34:1900-9. [Crossref] [PubMed]
- Kato T, Daigo Y, Aragaki M, et al. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol 2012;106:423-30. [Crossref] [PubMed]
- Durham KK, Chathely KM, Trigatti BL. High-density lipoprotein protects cardiomyocytes against necrosis induced by oxygen and glucose deprivation through SR-B1, PI3K, and AKT1 and 2. Biochem J 2018;475:1253-65. [Crossref] [PubMed]
- Mandeville I, Aubin J, LeBlanc M, et al. Impact of the loss of Hoxa5 function on lung alveogenesis. Am J Pathol 2006;169:1312-27. [Crossref] [PubMed]
- Alharbi RA, Pandha HS, Simpson GR, et al. Inhibition of HOX/PBX dimer formation leads to necroptosis in acute myeloid leukemia cells. Oncotarget 2017;8:89566-79. [Crossref] [PubMed]


