
KCTD12 is a prognostic marker of breast cancer and correlates with tumor immune cell infiltration
Introduction
Owing to the remarkable advancements in genomics, multiple genes involved in tumor physiological processes have been identified. Most of these genes are considered tumor suppressors or oncogenes based on their role in tumorigenesis (1,2). In our study, we focused on the clinical applicability of potassium channel tetramerization domain containing 12 (KCTD12) in breast cancer (BC). The complementary deoxyribonucleic acid (cDNA) which encodes KCTD12 in humans was first isolated by Resendes et al. in 2004 (3). As a member of receptor complexes, the primary function of KCTD12 is to react to gamma-aminobutyric acid-B (GABA-B), thereby inducing GABA-B receptor-activated K+ current desensitization (4,5). Oncological studies have provided evidence that KCTD12 is involved in cancer cell stemness (6). For instance, Li et al. reported that silencing of KCTD12 could enhance colorectal cancer cell stemness through the extracellular-signal-related kinase (ERK) pathway (7), and Shen et al. demonstrated that depressed KCTD12 expression enhanced the stemness of melanoma cells by interacting with cluster of differentiation 271 (CD271) (8). Meanwhile, the series of studies from Suehara et al. initially found that expression levels of KCTD12 were inversely correlated with metastasis and/or recurrence rate of gastrointestinal stromal tumor (9,10). They also developed a mouse monoclonal antibody against pfetin (protein encoded by KCTD12) and evaluated its validation (11,12). However, KCTD12’s role in tumorigenesis remains controversial. Although one study that demonstrated that increased KCTD12 expression promoted tumorigenesis by facilitating cells’ G2/M transition in cervical and lung cancer (13), other studies explained that overexpression of KCTD12 inhibited colorectal cancer (7) and cutaneous melanoma (8) growth in vivo. Thus far, few studies have explored the role of KCTD12 in BC.
BC is the most common cancer diagnosed among females and is the leading cause of female cancer death worldwide (14). In 2018, it was estimated that there were 2,088,849 newly diagnosed BC cases and 626,679 deaths due to BC globally. The original molecular classification of BC, including estrogen receptor (ER)-positive/luminal-like, basal-like [also known as triple-negative BC (TNBC)], Erb-B2-positive [also called human epidermal growth factor receptor 2 (HER2)], and normal breast, was proposed in 2000 (15), and was sequenced by dividing the ER-positive/luminal-like into luminal A and luminal B in 2001 (16). Traditionally, this molecular classification has driven prognostic predictions and adjuvant therapy recommendations for early-stage BC (17,18). However, with the enhanced insights into the molecular architecture of BC, it is now widely accepted that BC is a heterogeneous disease (19,20). The relatively basic measures of molecular phenotypes do not offer a comprehensive picture of the molecular architecture of BC and results in many patients ultimately being over- or undertreated. It is important to note that the response of BC to the recommended therapies was inadequate in some cases depending on the molecular subtype. Therefore, novel biomarkers that assist in predicting the clinical features and prognosis of BC are required. These biomarkers may also help to guide therapeutic decision-making.
In this study, we attempted to clarify the correlation that KCTD12 expression has with the clinical features of BC, overall survival (OS), and recurrence-free survival (RFS). The main pathways in BC that KCTD12 was involved in were identified. We found that KCTD12 expression was positively correlated with the expression of tumor-infiltrating lymphocytes (TILs), programed cell-death protein 1 (PD-1), and programed cell-death ligand 1 (PD-L1). Our results suggest that KCTD12 could be considered a prognostic indicator and underlying biomarker to predict patient response to PD-1 or PD-L1 inhibitor treatment.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2099).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene expression data of human BC
Ribonucleic acid-sequencing (RNA-Seq) filings along with corresponding clinical information of BC patients were retrieved from the publicly available Cancer Genome Atlas (TCGA) program databases (https://www.cancer.gov/tcga) in December 2019. In total, 1,053 cancer samples and 111 normal samples analyzed. The Cancer Cell Line Encyclopedia (CCLE) provided genomic data of human cancer models, messenger ribonucleic acid (mRNA) expression data, and gene methylation data. The KCTD12 expression data and methylation data of 50 BC cell lines were derived from the CCLE for correlation analysis (21).
Survival analysis performed by Kaplan-Meier and PrognoScan
Kaplan-Meier survival analysis of BC patients’ OS and RFS were performed online (kmplot.com/analysis/) (22). Patients were grouped by the median value of KCTD12 expression level. We used the PrognoScan database for meta-analysis of the prognostic value of KCTD12 as a complement (23). A P value <0.05 was considered statistically significant.
Gene set enrichment analysis (GSEA)
The GSEA software was utilized to examine the significantly different pathways between high and low KCTD12–expressing BC samples. Enriched terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway with corrected P value <0.05 and a false discovery rate value <0.25 were identified.
Correlation analysis with KCTD1 expression levels with TILs
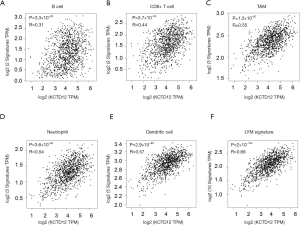
The Tumor Immune Estimation Resource (TIMER) was used to exam the correlation between KCTD12 expression and TILs or PD-1 and PD-L1 expression (24). The Gene Expression Profiling Interaction Analysis 2 (GEPIA2) was applied to investigate the correlation between KCTD12 expression and TILs markers. The gene markers of TILs including B cells, CD8+ (cytotoxic) T cells, tumor-associated macrophages (TAMs), neutrophils, and dendritic cells (DCs) are available in a previous study (25). In addition, the lymphocyte-specific immune recruitment (LYM) metagene signature is available in a previous study (26).
Statistical analysis
R (version 2.6.2) was utilized for statistical analysis. The Wilcoxon test was employed for differential analysis of KCTD12 expression in tumor and normal samples. The relationship between KCTD12 expression and the clinical features was analyzed using the Chi-Square test. Receiver operating characteristics (ROC) curves were generated by pROC package to assess the classification capability of KCTD12 and to set the optimal cutoff value to identify the high and low KCTD12 expression groups. Simple linear regression was used to evaluate the correlation between KCTD12 expression and methylation levels.
Results
KCTD12 was associated with BC clinical features
RNA-Seq data obtained from TCGA was analyzed to assess the relationship between KCTD12 expression levels and BC clinical features. Firstly, KCTD12 was significantly decreased in BC samples compared to unpaired normal samples (P=2.411×10−52) and paired normal samples (P=4.512×10−26) (Figure 1A,B). Secondly, the progesterone receptor (PR)-positive BC had a higher KCTD12 expression than that of PR-negative BC (P=0.00102) (Table 1). Thirdly, BC in small size (T1) and initial pathologic stages (Stage I) was accompanied by higher expression levels of KCTD12 in comparison with large size (T2–3) (P=0.00124) and advanced stages (Stage II–IV) BC (P=0.02025) (Table 1). Moreover, BC with low KCTD12 expression levels exhibited a short OS [hazard ratio (HR): 0.72 (0.58–0.9), P=0.003] and a short RFS [HR: 0.87 (0.78–0.97), P=0.012], indicating a poor prognosis (Figure 1C,D).
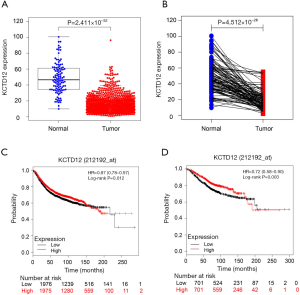
Table 1
| Features | Cases | KCTD12 in breast cancer (high/low expression), n | P value |
|---|---|---|---|
| Age at diagnosis, years | 0.11663 | ||
| ≤50 | 319 | 171/148 | |
| >50 | 716 | 346/370 | |
| ER status | 0.05389 | ||
| Positive | 765 | 399/366 | |
| Negative | 221 | 99/122 | |
| PR status | 0.00102 | ||
| Positive | 663 | 358/305 | |
| Negative | 320 | 137/183 | |
| HER2 status | 0.06133 | ||
| Positive | 173 | 77/96 | |
| Negative | 721 | 378/343 | |
| Pathologic stage | 0.02025 | ||
| I | 171 | 99/72 | |
| II/III/IV | 841 | 405/436 | |
| T stage | 0.00124 | ||
| 1 | 268 | 157/111 | |
| 2/3/4 | 764 | 360/404 | |
| N stage | 0.29537 | ||
| N0/N1 | 823 | 407/416 | |
| N2/N3 | 192 | 103/89 | |
| M stage | 0.11957 | ||
| M0 | 861 | 435/426 | |
| M1 | 21 | 7/14 |
ER, estrogen receptor; PR, progesterone receptor; T, tumor; N, lymph node; M, metastasis.
Also, the PrognoScan database provided multiple tests evaluating the association between KCTD12 expression and clinical outcome. Among these, 22 of the 55 tests showed that patients with decreased KCTD12 expression levels had significantly worse clinical outcomes than those with upregulated KCTD12 expression levels, while other tests showed no significant correlation between KCTD12 and prognosis. Representative tests are shown in Table S1. The above results demonstrated that KCTD12 was closely associated with patients’ clinical performance in BC.
In addition to BC, 11 other carcinoma types had significantly lower KCTD12 expression levels compared to normal tissues (Figure S1). We also compared the KCTD12 expression levels and methylation levels of CpG islands around the potassium channel tetramerization domain (KCTD) transcription start site in multiple BC cell lines. A medium reverse relation between KCTD12 expression and methylation levels was observed (Pearson’s coefficient = −0.45) (Figure S2).
KCTD12 exhibited the ability to reliably distinguish BC from normal tissue
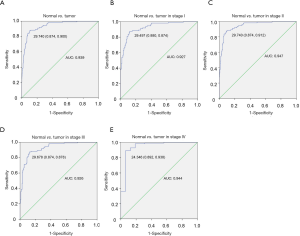
In addition to testing the correlation between KCTD12 and BC clinical performance, we also evaluated the efficiency of KCTD12 expression in distinguishing BC from normal breast tissues. As shown in Figure 2A, the area under the ROC (AUC) for distinguishing BC from normal tissues is 0.939 and the optimal cutoff value is annotated (Figure 2A). Across stages, the classification based on KCTD12 expression also has strong efficiency (Figure 2B,C,D,E).
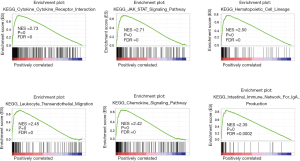
GSEA revealed that the KCTD12 participated in immune response in BC
To identify the main biological functions of KCTD12 in BC, GSEA was applied on 1,053 BC samples grouped by KCTD12 expression levels. We found that 6 out of the 10 top-ranked pathways were immune-associated, including cytokine-cytokine receptor interaction, Janus kinase (JAK)-signal transducer and activator transcription (STAT) signaling pathway, hematopoietic cell lineage, leukocyte transendothelial migration, chemokine signaling pathway, and intestinal immune network for immunoglobulin A (IgA) production (Figure 3). Similar results were observed when the GSEA was performed on 51 BC cell lines grouped by KCTD12 expression. The majority of the top 10 ranked pathways that KCTD12 participated in were also immune-associated (Figure S3). According to these results, it is highly probable that KCTD12 plays a role in the immune response in BC. Therefore, in subsequent analysis, we focused on the correlation between KCTD12 and tumor immune microenvironment.
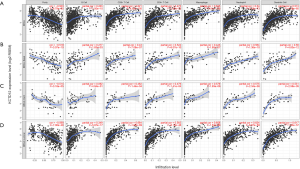
BC that highly expressed KCTD12 was accompanied with increased TILs
Previous studies have confirmed the high prognostic value of TILs in multiple cancers, including BC (27-30). We utilized the TIMER to evaluate the relationship between KCTD12 and TILs [including B cells, CD4+ (helper) T cells, CD8+ T cells, neutrophil, macrophage, and dendritic cells]. As shown in Figure 4, KCTD12 expression levels are positively correlated with an abundance of all six types of TILs in BC. This correlation was present across all BC subtypes and TIL types, expect for the B cell in HER-2-positive BC. These results demonstrate that KCTD12 expression can reflect the abundance of TILs across all subtypes of BC.
KCTD12 was associated with increased markers of TILs
We further used the GEPIA2 to confirm that our results concerning the relationship between KCTD12 mRNA levels and BC TILs were true, independent of the analysis method used. The results demonstrated that KCTD12 mRNA quantities were positively correlated with markers of several lymphocytes, including B cell, CD8+ cell, TAM, neutrophil, and DC (Figure 5A,B,C,D,E). Moreover, as a prognosis metagene signature of BC, the LYM indicated the immune phenotype of BC (26). So, we evaluated the correlation between the expression levels of KCTD12 and LYM and found that KCTD12 expression levels were positively correlated with LYM expression levels (Figure 5F). Combined with results we obtained from the TIMER, we concluded that elevated KCTD12 expression levels in BC reflected significant lymphocyte infiltration.
KCTD12 expression levels were positively correlated to PD-1 and PD-L1 expression levels
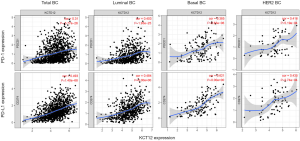
Immune checkpoint blockade (ICB) agents have received increasing attention, as they have been shown to provide durable responses and prolong OS in several solid tumors (31-34). The cytotoxic T-lymphocyte antigen 4 (CTLA-4), PD-1, and PD-L1 are considered main immune checkpoint proteins. As a representative of ICB agents, the anti-PD-1 antibody, atezolizumab, has received United States Food and Drug Administration (FDA) approval to be applied for metastatic triple-negative breast cancer (TNBC) in combination with nab-paclitaxel (35,36).
We subsequently tested the correlation between KCTD12 expression and PD-1 and PD-L1 expression using the TIMER. We observed that KCTD12 expression levels were positively correlated with both PD-1 and PD-L1 expressions, either in all BC or across subtypes of BC (Figure 6). Therefore, this demonstrated that KCTD12 was positively associated with immune checkpoint expression. The higher KCTD12 expression in BC may suggest a higher response rate to PD-1 and PD-L1 inhibitors.
Discussion
KCTD12 has been studied extensively in gastrointestinal stromal tumor and is reportedly involved in the physiological processes of several types of carcinomas (9-12). Yet, related studies in BC are rare (37). In this study, we found that KCTD12 was closely associated with the clinical features of BC. KCTD12 expression was decreased in BC and 11 other types of carcinomas compared to corresponding normal tissues. The lower expression levels in advanced stages indicated that KCTD12 was decreased in aggressive disease. Conversely, BC with higher KCTD12 expression had a more favorable prognosis in Kaplan-Meier survival analysis and PrognoSan database analysis, indicating that KCTD12 has prognostic value in BC. A similar relation between KCTD12 expression and favorable prognosis was also observed in gastrointestinal stromal tumor (10), colorectal cancer (7), and cutaneous melanoma (8). According to these results, and together with a study reporting that the downregulation of KCTD12 significantly promoted BC cell proliferation (37), we suggest that loss of function of KCTD12 may contribute to BC tumorigenesis. Meanwhile, an inverse relationship between KCTD12 expression levels and methylation levels was observed in BC cell lines, which indicates the possibility that the hypermethylation of the KCTD12 promoter leads to decreased mRNA levels.
GSEA revealed a previously unreported function of KCTD12 in carcinoma, namely immune response. Traditionally, based on the tumor microenvironment, tumors can be characterized as either “hot” (containing enriched TILs) or “cold” (containing few TILs) tumors (38). “Hot” BC is predicted to have better outcomes when chemotherapy, radiation therapy, or immunotherapy are applied. For example, patients with non-metastatic, basal-like cancer containing enriched TILs have improved outcomes compared to those containing poor TILs when treated with standard therapeutic regimens (27,39). Patients with HER-2-positive BC also experience better treatment outcomes with anti-HER2 antibody trastuzumab when the tumor has significant lymphocyte infiltration (40). Here, we evaluated the relationship between KCTD12 and TILs in BC. The positive correlation between KCTD12 expression and TILs was observed across all BC subtypes. In addition, the KCTD12 was shown to be correlated to PD-L1 and PD-1 expression in BC.
As a new and promising cancer treatment, ICB therapy receives increasing investment in the treatment of solid tumors (35), and the biomarkers that predict the reaction of BC to ICB agents also deserve attention. Atezolizumab, an anti-PD-L1 antibody, has been the only ICB agent approved by the FDA for clinical treatment of BC thus far (41). Meanwhile, the PD-L1 expression analyzed by immunohistochemistry (IHC) has been shown to be an efficient biomarker for selecting patients who would benefit from anti-PD-1 and anti-PD-L1 antibodies (42,43). In fact, TIL quantities and PD-L1 expression have been embedded in multifactorial biomarkers of clinical response to anti-PD-1 and anti-PD-L1 antibodies. The positive correlation of KCTD12 to TIL abundance and PD-L1 and PD-1 expression indicates that it could be a predictive marker to identify BC patients who may benefit more from anti-PD-1 and anti-PD-L1 antibodies. Put simply, the expression of KCTD12 can predict prognosis and may also help to estimate the response of patients treated with ICB agents.
Conclusions
Our study demonstrated that KCTD12 expression is closely related to the clinical features of BC, with high KCTD12 expression levels in BC being associated with a favorable prognosis. KCTD12 expression also proved to be positively connected to TIL quantities and PD-L1 and PD-1 expression, suggesting that KCTD12 could be considered a potential biomarker to identify those BC patients who may derive greater benefit from anti-PD-L1 and anti-PD-1 antibodies.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2099
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2099). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee EYHP, Muller WJ. Oncogenes and Tumor Suppressor Genes. Cold Spring Harb Perspect Biol 2010;2:a003236. [Crossref] [PubMed]
- Lane DP. p53, guardian of the genome. Nature 1992;358:15-6. [Crossref] [PubMed]
- Resendes BL, Kuo SF, Robertson NG, et al. Isolation from cochlea of a novel human intronless gene with predominant fetal expression. J Assoc Res Otolaryngol 2004;5:185-202. [Crossref] [PubMed]
- Schwenk J, Metz M, Zolles G, et al. Native GABAB receptors are heteromultimers with a family of auxiliary subunits. Nature 2010;465:231-5. [Crossref] [PubMed]
- Turecek R, Schwenk J, Fritzius T, et al. Auxiliary GABAB Receptor Subunits Uncouple G Protein βγ Subunits from Effector Channels to Induce Desensitization. Neuron 2014;82:1032-44. [Crossref] [PubMed]
- Abbaszadegan MR, Taghehchian N, Li L, et al. Contribution of KCTD12 to esophageal squamous cell carcinoma. BMC Cancer 2018;18:853. [Crossref] [PubMed]
- Li L, Duan T, Wang X, et al. KCTD12 Regulates Colorectal Cancer Cell Stemness through the ERK Pathway. Sci Rep 2016;6:20460. [Crossref] [PubMed]
- Shen W, Li Y, Li B, et al. Downregulation of KCTD12 contributes to melanoma stemness by modulating CD271. Cancer Biol Med 2019;16:498-513. [PubMed]
- Suehara Y, Kondo T, Seki K, et al. Pfetin as a Prognostic Biomarker of Gastrointestinal Stromal Tumors Revealed by Proteomics. Clin Cancer Res 2008;14:1707-17. [Crossref] [PubMed]
- Kubota D, Orita H, Yoshida A, et al. Pfetin as a Prognostic Biomarker for Gastrointestinal Stromal Tumor: Validation Study in Multiple Clinical Facilities. Jpn J Clin Oncol 2011;41:1194-202. [Crossref] [PubMed]
- Kubota D, Mukaihara K, Yoshida A, et al. The Prognostic Value of Pfetin: a Validation Study in Gastrointestinal Stromal Tumors Using a Commercially Available Antibody. Jpn J Clin Oncol 2013;43:669-75. [Crossref] [PubMed]
- Kikuta K, Gotoh M, Kanda T, et al. Pfetin as a Prognostic Biomarker in Gastrointestinal Stromal Tumor: Novel Monoclonal Antibody and External Validation Study in Multiple Clinical Facilities. Jpn J Clin Oncol 2010;40:60-72. [Crossref] [PubMed]
- Zhong Y, Yang J, Xu WW, et al. KCTD12 promotes tumorigenesis by facilitating CDC25B/CDK1/Aurora A-dependent G2/M transition. Oncogene 2017;36:6177-89. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 2001;98:10869-74. [Crossref] [PubMed]
- Cianfrocca M, Gradishar W. New Molecular Classifications of Breast Cancer. CA Cancer J Clin 2009;59:303-313. [Crossref] [PubMed]
- Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. The Breast 2015;24:S26-35. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2009;2:18. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: a Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017;12:e0179726. [Crossref] [PubMed]
- Cheng WY, Yang THO, Anastassiou D. Development of a Prognostic Model for Breast Cancer Survival in an Open Challenge Environment. Sci Transl Med 2013;5:181ra50. [Crossref] [PubMed]
- Loi S, Sirtaine N, Piette F, et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. J Clin Oncol 2013;31:860-7. [Crossref] [PubMed]
- Dieci MV, Mathieu MC, Guarneri V, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann Oncol 2015;26:1698-704. [Crossref] [PubMed]
- Adams S, Gray RJ, Demaria S, et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancers From Two Phase III Randomized Adjuvant Breast Cancer Trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014;32:2959-66. [Crossref] [PubMed]
- Ibrahim EM, Al-Foheidi ME, Al-Mansour MM, et al. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: a meta-analysis. Breast Cancer Res Treat 2014;148:467-76. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Hamid O, Robert C, Daud A, et al. Safety and Tumor Responses with Lambrolizumab (Anti-PD-1) in Melanoma. N Engl J Med 2013;369:134-44. [Crossref] [PubMed]
- Robert C, Schachter J, Long G V, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 2015;372:2521-32. [Crossref] [PubMed]
- Adams S, Gatti-Mays ME, Kalinsky K, et al. Current Landscape of Immunotherapy in Breast Cancer. JAMA Oncol 2019;5:1205. [Crossref] [PubMed]
- Esteva FJ, Hubbard-Lucey VM, Tang J, et al. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol 2019;20:e175-86. [Crossref] [PubMed]
- Ye RY, Kuang XY, Zeng HJ, et al. KCTD12 promotes G1/S transition of breast cancer cell through activating the AKT/FOXO1 signaling. J Clin Lab Anal 2020;34:e23315. [Crossref] [PubMed]
- Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674-90. [Crossref] [PubMed]
- Savas P, Salgado R, Denkert C, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol 2016;13:228-41. [Crossref] [PubMed]
- Loi S, Michiels S, Salgado R, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol 2014;25:1544-50. [Crossref] [PubMed]
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350-5. [Crossref] [PubMed]
- Emens LA, Cruz C, Eder JP, et al. Long-term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients With Metastatic Triple-Negative Breast Cancer. JAMA Oncol 2019;5:74. [Crossref] [PubMed]
- Dirix LY, Takacs I, Jerusalem G, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat 2018;167:671-86. [Crossref] [PubMed]
(English Language Editors: A. Kassem and J. Gray)


