
A novel nomogram for predicting survival of patients with poorly differentiated gastric adenocarcinoma
Introduction
Gastric cancer (GC) is the third cause of cancer-related deaths, and a significant threat to human health worldwide (1). GC is a heterogeneous disease with various histological and molecular subtypes (2). In fact, recent research progress has revealed four molecular GC subtypes, including Epstein-Barr virus (EBV)-positive, the microsatellite instability (MSI), chromosome instability (CIN), and genomically stable (GS) subtype (3). Accurate diagnosis and development of effective treatment therapies require an effective way to simplify disease’s heterogeneity based on its subtypes to study (4). Poorly differentiated gastric adenocarcinoma (PDGA), a common pathological type of gastric cancer, is an adenocarcinoma with less glandular structure (5). According to classifications by the World Health Organization (WHO) (6) and the Japanese classification of gastric carcinoma (7), poorly differentiated adenocarcinoma (PDA) comprises tumors that exhibit various morphologies, such as adenocarcinoma with poor cohesion, signet ring cell carcinoma, among others (5). There was a previous study showing that the PDGA showed weaker expression of CPP32, EMMPRIN, MUC-2, MUC-5AC, and MUC-6 (8). Yang et al. also reported that Epstein-Barr virus associated gastric cancer (EBVaGC) was positively associated with PDGA (9). In the present study, we focused on four common histological types of PDA in gastric cancer, including poorly differentiated adenocarcinoma, undifferentiated adenocarcinoma, signet ring cell carcinoma, and mucinous adenocarcinoma.
Currently, the therapies for treating PDGA include surgical resection, radiotherapy, chemotherapy and molecular targeted therapy (10). Although significant progress has been made on this front, prognosis of PDGA remains challenging due to recurrence and metastasis (11,12). Furthermore, several studies have reported inconsistent results with regards to differentiation status, possibly due to varying sample sizes and inclusion criteria (13-16). Numerous studies have reported occurrence of MSI in solid-type PDA of the stomach. Particularly, MSI-positive solid PDA is more common in older, female gastric cancer patients, with pathological characteristics of these patients revealing consistent glandular composition and a significant correlation between MLH1 and PMS2 expression loss (5). One study has also demonstrated that solid-type PDA frequently lose MMR protein and the SWI/SNF complex (17). However, most patients with differentiated gastric adenocarcinoma tend to be male, with the tumors generally appearing hematogenous (18). Based on these, we hypothesized that the underlying prognostic factors for PDGA may be different from other histological types of gastric cancer. Therefore, developing a more specific therapy that directly targets PDGA may have clinical value in guiding development of treatment therapies. Consequently, we sought to explore the clinicopathological features of PDGA and formulate more detailed and effective treatment plans.
Although there were several reports about nomogram for predicting survival of patients with gastric neuroendocrine neoplasms (19,20), up to date, no studies have separately reported the specific prognostic factors of PDGA. In view of the fact that histological type is essential to assess the tumor progression and prognosis (21), and PDGA is a common histopathological type in gastric cancer, we believe that it is necessary to establish a nomogram to predict overall survival (OS) for PDGA patients. Specifically, we targeted the Surveillance, Epidemiology, and End Results (SEER) program of the National Cancer Institute (NCI), which collects data on approximately 450,000 cases of malignant and in situ cancers each year in the United States. This database provides data on patient demographics, tumor morphology, stage at diagnosis, primary tumor sites, survival times, and vital status (22). In our study, we retrospectively analyzed clinicopathological features of PDGA patients based on the larger sample size in the SEER database, screened out independent prognostic risk factors, then used them to develop a nomogram for predicting prognosis of PDGA patients. In addition, we compared the prognostic power of our nomogram to the 8th edition of the American Joint Committee on Cancer (AJCC) tumor TNM staging system, and finally validated it externally using data from an independent Chinese set.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2794).
Methods
Patient enrolment and data retrieval
We screened the SEER database (http://seer.cancer.gov/) and retrieved data from gastric cancer patients diagnosed with pathological PDA and who underwent surgery between 2010 and 2015 at Zhejiang Provincial People’s Hospital. The last follow-up date of validation sets was January 2018. We extracted cases of gastric cancer patients, with histological type PDA, based on the International Classification of Diseases for Oncology (ICD-O-3), 3rd edition. We further retrieved demographic and clinical information including age, gender, histological differentiation type, grade, primary tumor site, tumor size, tumor number, and the 7th AJCC TNM stage. We redefined the TNM staging according to the criterion described in the 8th AJCC guidelines. The inclusion criteria were as follows: (I) gastric cancer patients who underwent surgery; and (II) ICD-O-3 codes for histological type were adenocarcinomas [8140-8389]; (III) poorly differentiated or undifferentiated/anaplastic histological grade III or grade IV, respectively. Conversely, the exclusion criteria included: (I) without complete follow-up information; (II) without detail TNM stage information; (III) unknown histological differentiation type; (IV) unknown grade; (V)unknown primary tumor site; (VI) unknown tumor size; and (VII) unknown tumor number. In this study, the training set’s data comes from a public database (SEER database) which does not require ethical approval. However, the validation set data from Zhejiang Provincial People’s Hospital requires an ethics statement. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Zhejiang Provincial People’s Hospital (Approval No. 2019KY017) and informed consent was taken from all the patients.
Construction of the nomogram
We used the retrieved SEER dataset as a training set to build an OS prognostic nomogram. First, we performed univariate analysis to screen for significant variables related to prognosis (P<0.05). Furthermore, we used multivariate Cox proportional hazard regression analysis to identify independent risk factors related to prognosis, then employed them to construct a prognostic nomogram using the rms package implemented in R software (“http://www.r-project.org/”). The resultant nomogram was subsequently used to predict 1-, 3- and 5-year OS for gastric cancer patients with PDA.
Nomogram validation
An independent set of Chinese patient data was used to externally validate prognostic performance of the nomogram. Briefly, we evaluated the discriminative power of our nomogram using the concordance index (C-index), by measuring the difference in predictive power between observed and the predicted results. In addition, we plotted receiver operating characteristic (ROC) curves and calculated the area under curve (AUC) to evaluate accuracy of the 1-, 3-and 5-year survival predictions.
Statistical analysis
We used the chi-square test to compare variables between training and validation sets, then performed survival analysis using Kaplan-Meier method and the log-rank test. Furthermore, we assessed the effectiveness of our nomogram to predict prognosis of patients by generating C index and AUC, then compared the results with those from the TNM staging (8th AJCC). All statistical analyses were performed using packages implemented in R software version 3.5.2 (R foundation for Statistical Computing, Vienna, Austria) at a 95% confidence interval (CI). Data followed by P<0.05 were considered statistically significant.
Results
Patient characteristics
Screening the SEER database revealed a total of 3,887 patients who met our inclusion criteria, and were therefore included in the training set. On the other hand, 632 Chinese patients met these criteria and were subsequently enrolled as a validation set (Figure 1). A summary of the patients’ clinical characteristics across both study sets is outlined in (Table 1). In the SEER training set, 2,525 (64.96%) and 1,362 (35.04%) of the patients were males and females, respectively. Their median ages were 68 years (range, 19–97 years) and 34 months at diagnosis and follow-up, respectively. In the Chinese validation set, there were 423 (66.93%) and 209 (33.07%) males and females. Respectively, with median ages of 63 years (range, 21–89 years) and 31 months at diagnosis and follow-up, respectively. The proportions of tumor size ≤6.8 cm for the training and validation sets were 79.58 and 81.49%, respectively. In addition, patients in the SEER set exhibited even distribution of primary tumor sites in the stomach. Conversely, the gastric antrum or pylorus, accounted for the vast majority (71.99%) of the tumor site in the Chinese set. A higher proportion of single tumors was recorded in the Chinese, than the SEER set, at 98.26 and 78.67%, respectively. Moreover, a higher proportion of patients in the Chinese set (52.85%) who underwent surgery were diagnosed with stage III cancer. On the other hand, we found no significant differences between the groups with regards to sex, tumor size, and liver metastasis, although age, prime site of tumor, tumor number, and TNM stage (8th AJCC) were significantly different (P<0.05).
Table 1
| Demographic or characteristics | SEER training set (n=3,887) | Chinese validation set (n=632) | P value | |||
|---|---|---|---|---|---|---|
| No. | Percentage, % | No. | Percentage, % | |||
| Age at diagnosis (year) | <0.001 | |||||
| ≤44 | 220 | 5.66 | 47 | 7.44 | ||
| 45–59 | 863 | 22.20 | 193 | 30.54 | ||
| 60–74 | 1,642 | 42.24 | 280 | 44.30 | ||
| ≥75 | 1,162 | 29.89 | 112 | 17.72 | ||
| Sex | 0.335 | |||||
| Female | 1,362 | 35.04 | 209 | 33.07 | ||
| Male | 2,525 | 64.96 | 423 | 66.93 | ||
| Tumor size (cm) | 0.436 | |||||
| <3.7 | 1,555 | 40.01 | 251 | 39.72 | ||
| 3.7–6.8 | 1,538 | 39.57 | 264 | 41.77 | ||
| >6.8 | 794 | 20.43 | 117 | 18.51 | ||
| Prime site of tumor | <0.001 | |||||
| Cardia/fundus | 1,290 | 33.19 | 131 | 20.73 | ||
| Middle of gastric body* | 1,222 | 31.44 | 46 | 7.28 | ||
| Antrum/pylorus | 1,375 | 35.37 | 455 | 71.99 | ||
| Tumor number | <0.001 | |||||
| <2 | 3,058 | 78.67 | 621 | 98.26 | ||
| ≥2 | 829 | 21.33 | 11 | 1.74 | ||
| AJCC T stage (8th) | <0.001 | |||||
| T1 | 612 | 15.74 | 66 | 10.44 | ||
| T2 | 500 | 12.86 | 110 | 17.41 | ||
| T3 | 1,733 | 44.58 | 234 | 37.03 | ||
| T4 | 1,042 | 26.81 | 222 | 35.13 | ||
| AJCC N stage (8th) | <0.001 | |||||
| N0 | 1,420 | 36.53 | 197 | 31.17 | ||
| N1 | 808 | 20.79 | 96 | 15.19 | ||
| N2 | 774 | 19.91 | 115 | 18.20 | ||
| N3a | 631 | 16.23 | 147 | 23.26 | ||
| N3b | 254 | 6.53 | 77 | 12.18 | ||
| AJCC M stage (8th) | <0.001 | |||||
| M0 | 3,536 | 90.97 | 606 | 95.89 | ||
| M1 | 351 | 9.03 | 26 | 4.11 | ||
| AJCC TNM stage (8th) | <0.001 | |||||
| I | 790 | 20.32 | 134 | 21.20 | ||
| II | 1,255 | 32.29 | 138 | 21.84 | ||
| III | 1,491 | 38.36 | 334 | 52.85 | ||
| IV | 351 | 9.03 | 26 | 4.11 | ||
| Liver metastasis | 0.354 | |||||
| No | 3,761 | 96.76 | 607 | 96.04 | ||
| Yes | 126 | 3.24 | 25 | 3.96 | ||
*, middle of gastric body includes body of stomach, lesser curvature of stomach, and greater curvature of stomach. T, N, M stages come from the 8 edition AJCC Staging. AJCC, American Joint Committee on Cancer; SEER, Surveillance, Epidemiology, and End Results.
Nomogram establishment
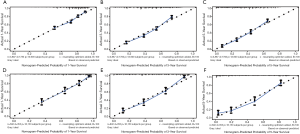
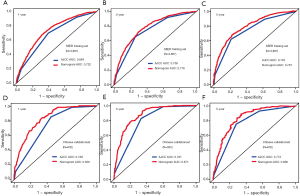
Univariate analysis of the collected factors in the SEER training set revealed seven significant prognostic factors, including age, sex, tumor size, prime site of tumor, T stage (8th AJCC), N stage (8th AJCC), and M stage (8th AJCC) (Table 2). Multivariate Cox analysis confirmed that these 7 factors were indeed independent prognostic risk factors for gastric cancer patients with PDA (Table 3). Since the TNM stage interacts with other factors, we excluded it from the Cox-regression model to prevent multicollinearity that renders model estimation thereby compromising accuracy. Subsequently, we used the selected independent predictive factors to establish a nomogram for 1-, 3- and 5-year OS (Figure 2). Summarily, the nomogram predicts the 1-, 3- and 5-year OS for each patient by summing up the scores displayed on the bottom scale. The calibration plots revealed an optimal agreement between nomogram-predicted OS and actual ones estimated by the Kaplan-Meier method (Figure 3). In addition, our nomogram’s OS C-index was 0.707, whereas its AUCs for predicting the 1-, 3- and 5-year OS were 0.732, 0.776 and 0.787, respectively (Figure 4A,B,C).
Table 2
| Variables | Univariate analysis | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (year) | ||
| ≤44 | Reference | |
| 45–59 | 1.072 (0.119–0.588) | 0.557 |
| 60–74 | 1.315 (0.114–2.408) | 0.016 |
| ≥75 | 2.178 (0.114–6.797) | <0.001 |
| Sex | ||
| Female | Reference | |
| Male | 1.118 (0.052–2.143) | 0.032 |
| Tumor size (cm) | ||
| <3.7 | Reference | |
| 3.7–6.8 | 1.136 (0.059–2.183) | 0.029 |
| >6.8 | 1.105 (0.069–1.442) | 0.149 |
| Prime site of tumor | ||
| Cardia/fundus | Reference | |
| Middle of gastric bodya | 0.694 (0.063–5.814) | <0.001 |
| Antrum/pylorus | 0.768 (0.061–4.338) | <0.001 |
| Tumor number | ||
| <2 | Reference | |
| ≥2 | 1.039 (0.058–0.669) | 0.504 |
| AJCC T stage (8th) | ||
| T1 | Reference | |
| T2 | 1.153 (0.118–1.209) | 0.227 |
| T3 | 1.685 (0.098–5.335) | <0.001 |
| T4 | 2.380 (0.105–8.239) | <0.001 |
| AJCC N stage (8th) | ||
| N0 | Reference | |
| N1 | 1.704 (0.074–7.161) | <0.001 |
| N2 | 2.091 (0.075–9.888) | <0.001 |
| N3a | 2.657 (0.078–12.546) | <0.001 |
| N3b | 3.209 (0.098–11.853) | <0.001 |
| AJCC M stage (8th) | ||
| M0 | Reference | |
| M1 | 1.993 (0.086–8.039) | <0.001 |
| AJCC TNM stage (8th) | ||
| I | Reference | |
| II | – | <0.001 |
| III | – | <0.001 |
| IV | – | <0.001 |
| Liver metastasis | ||
| No | Reference | |
| Yes | 1.221 (0.128–1.568) | 0.117 |
a, middle of gastric body includes body of stomach, lesser curvature of stomach, and greater curvature of stomach. T, N, M stages come from AJCC. AJCC, American Joint Committee on Cancer; SEER, Surveillance, Epidemiology, and End Results.
Table 3
| Variables | Multivariate analysis | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤44 | Reference | |
| 45–59 | 1.078 (0.118–0.635) | 0.525 |
| 60–74 | 1.331 (0.113–2.531) | 0.011 |
| ≥75 | 2.215 (0.113–7.021) | <0.001 |
| Sex | ||
| Female | Reference | |
| Male | 1.119 (0.052–2.170) | 0.030 |
| Tumor size (cm) | ||
| <3.7 | Reference | |
| 3.7–6.8 | 1.137 (0.059–2.193) | 0.028 |
| >6.8 | 1.110 (0.069–1.512) | 0.131 |
| Prime site of tumor | ||
| Cardia/fundus | Reference | |
| Middle of gastric bodya | 0.689 (0.063–5.935) | <0.001 |
| Antrum/pylorus | 0.762 (0.061–4.451) | <0.001 |
| AJCC T stage (8th) | ||
| T1 | Reference | |
| T2 | 1.152 (0.118–1.203) | 0.229 |
| T3 | 1.677 (0.098–5.288) | <0.001 |
| T4 | 2.363 (0.105–8.172) | <0.001 |
| AJCC N stage (8th) | ||
| N0 | Reference | |
| N1 | 1.709 (0.074–7.206) | <0.001 |
| N2 | 2.090 (0.075–9.882) | <0.001 |
| N3a | 2.661 (0.078–12.575) | <0.001 |
| N3b | 3.210 (0.098–11.870) | <0.001 |
| AJCC M stage (8th) | ||
| M0 | Reference | |
| M1 | 2.139 (0.071–10.784) | <0.001 |
a, middle of gastric body includes body of stomach, lesser curvature of stomach, and greater curvature of stomach. T, N, M stages come from AJCC. AJCC, American Joint Committee on Cancer; SEER, Surveillance, Epidemiology, and End Results.
External nomogram validation and assessment of its efficiency
External validation, using data from 632 Chinese patients from Zhejiang Provincial People’s Hospital, confirmed the ability of our nomogram to predict OS. Specifically, the C-index for evaluating accuracy of the nomogram was 0.788, while AUC values for predicting 1-, 3-, and 5-year OS of the nomogram found to be 0.826, 0.871, and 0.836, respectively (Figure 4D,E,F). In SEER training set, the C-index of the 8th AJCC TNM staging system was 0.663, whereas AUC values of the ROC for predicting 1-, 3-, and 5-year OS were 0.684, 0.738, and 0.745, respectively (Figure 4A,B,C). In the Chinese validation set, the C-index of the 8th AJCC TNM staging system was 0.713, whereas AUC values of the ROC for predicting 1-, 3-, and 5-year OS were 0.738, 0.791, and 0.773, respectively (Figure 4D,E,F). Overall, our nomogram showed higher prognostic accuracy than the 8th AJCC TNM staging system across the training and validation sets (Table 4).
Table 4
| System | C-index | AUC | ||
|---|---|---|---|---|
| 1-year survival | 3-year survival | 5-year survival | ||
| SEER training set | ||||
| The 8th AJCC TNM Staging System | 0.663 | 0.684 | 0.738 | 0.745 |
| Nomogram | 0.707 | 0.732 | 0.776 | 0.787 |
| P value | <0.001 | <0.001 | <0.001 | <0.001 |
| Chinese validation set | ||||
| The 8th AJCC TNM Staging System | 0.713 | 0.738 | 0.791 | 0.773 |
| Nomogram | 0.788 | 0.826 | 0.871 | 0.836 |
| P value | <0.001 | <0.001 | <0.001 | <0.001 |
AUC, area under the curve; AJCC, American Joint Committee on Cancer; SEER, Surveillance, Epidemiology, and End Results.
Discussion
Individualized gastric cancer treatment, a multi-disciplinary collaboration and supplementation approach projected to improve efficacy of cancer treatment, is currently the focus of numerous medical research. Since accurate tumor typing forms the basis of individualized tumor treatment, it is imperative to ensure proper classification of GC (23). Previous studies have described the importance of histological type in evaluating tumor progression and prognosis of gastric cancer patients (21). Furthermore, PDA, a common histopathological type of gastric cancer, has been implicated in poor prognosis of PDA than other types, although some studies have shown that PDA with good morphology has a good prognosis (5,17,18). In the present study, we believe that exploring prognostic risk factors in PDA patients is important, owing to its high incidence in gastric cancer. From our analyses, it was evident that some differences existed between PDGA patients in the SEER database and Chinese PDGA patients. Specifically, we found statistically significant differences between the groups with regards to clinicopathological characteristics, including age at diagnosis, prime site of tumor, tumor number, AJCC T stage (8th), AJCC N stage (8th), and AJCC M stage (8th) in the two sets, which may result from a complex interaction between race, geographic location, culture, eating habits, and socioeconomic inequality. In addition, univariate a multivariate analysis of the clinicopathological characteristics and independent prognostic factors of 3,887 gastric cancer patients in the SEER database revealed 7 independent risk factors related to OS. These factors, including age, sex, tumor size, prime site of tumor, T stage (8th AJCC), T stage (8th AJCC), and T stage (8th AJCC) were used to build an OS nomogram model, which was externally validated using a Chinese patient population. The larger sample size of the two sets of data improves the accuracy and credibility of the research. And we used the ROC curve (1, 3, and 5 years of AUC value), the calibration plots, the C index (the value of the C index) to evaluate the accuracy of the model, and found that the prognostic model has high accuracy.
The AJCC TNM staging system for malignant tumors has long been used as a basis for clinical treatment (24). However, response to treatment and prognosis are not usually consistent if the same treatment plan is adopted in patients with tumors with the same TNM stage (25-27). Although the TNM staging system is based on data collected globally, data from two countries, South Korea and Japan, account for 84.8% of data from Asian countries, which may have a significant impact on the analysis of Chinese patients (28). Fortunately, our OS nomogram model maybe solve this problem (29,30). Similarly, our nomogram was more accurate at predicting the OS of PDGA patients than the AJCC-TNM staging system, across both training and validation sets (C-index value in the SEER training set: 0.707 vs. 0.663; C-index value in the Chinese validation set: 0.788 vs. 0.713). In addition, the nomogram had a more prominent advantage than the 8th edition of the AJCC staging system, in that it integrated more potential independent prognostic risk factors to make personalized predictions of patient survival, thereby making better treatment strategies. Despite some differences between the Chinese and SEER sets, our nomogram still showed acceptable consistency in the external verification set, indicating that these differences will not reduce the effectiveness of the nomogram. Moreover, the calibration chart revealed excellent agreement between predicted probability and the actual observation, thereby affirming reliability and repeatability of the nomogram constructed herein.
Our study had several limitations. Firstly, we did not include racial factors, which may affect the accuracy of our results. Although there are differences in diet and treatment between American and Chinese patients, some basic information between patients is still comparable, such as the TNM staging of tumors. And there is currently no large public gastric cancer database available for analysis in China. Therefore, we used the huge data of the SEER database to establish a nomogram and verified it with 632 samples collected from a single center in China. Therefore, this research still has certain reference value. Secondly, our external validation only used data collected from a single-center PDGA patient in China. Thirdly, treatment bias may have occurred. Since some patients in the SEER database had incomplete chemotherapy or radiotherapy data, or recorded data on chemotherapy or radiotherapy, but did not specify the specific chemotherapy or radiotherapy program. Considering that different chemotherapy or radiotherapy regimens may have different effects on the prognosis of patients with gastric cancer, and some chemotherapy or radiotherapy data are incomplete, this study did not include chemotherapy or radiotherapy data, which is also the focus of our future research.
Conclusions
In conclusion, we successfully established a novel nomogram using patient data from the SEER database, and validated its efficiency in predicting prognosis of PDGA patients using a Chinese population. Our nomogram had more accurate and effective than the 8th AJCC TNM staging system, hence more suitable for future development of personalized treatment for patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist Available at http://dx.doi.org/10.21037/tcr-20-2794
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2794
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2794). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Zhejiang Provincial People’s Hospital (Approval No.: 2019KY017) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Yan HHN, Siu HC, Law S, et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018;23:882-97.e11. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Lim B, Kim JH, Kim M, et al. Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J Gastroenterol 2016;22:1190-201. [Crossref] [PubMed]
- Arai T, Matsuda Y, Aida J, et al. Solid-type poorly differentiated adenocarcinoma of the stomach: clinicopathological and molecular characteristics and histogenesis. Gastric Cancer 2019;22:314-22. [Crossref] [PubMed]
- Fléjou JF. WHO Classification of digestive tumors: the fourth edition. Ann Pathol 2011;31:S27-31.
- Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer 2011;14:101-12. [Crossref] [PubMed]
- Zheng HC, Zheng YS, Xia P, et al. The pathobiological behaviors and prognosis associated with Japanese gastric adenocarcinomas of pure WHO histological subtypes. Histol Histopathol 2010;25:445-52. [PubMed]
- Yang Y, Liu YQ, Wang XH, et al. Clinicopathological and molecular characteristics of Epstein-Barr virus associated gastric cancer: a single center large sample case investigation. Beijing Da Xue Xue Bao Yi Xue Ban 2019;51:451-8. [PubMed]
- Oshima T, Masuda M. Molecular targeted agents for gastric and gastroesophageal junction cancer. Surg Today 2012;42:313-27. [Crossref] [PubMed]
- Dong D, Fang MJ, Tang L, et al. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multi-center study. Ann Oncol 2020;31:912-20. [Crossref] [PubMed]
- Fujimoto A, Ishikawa Y, Ishii T, et al. Differences between gastric signet-ring cell carcinoma and poorly differentiated adenocarcinoma: A comparison of histopathologic features determined by mucin core protein and trefoil factor family peptide immunohistochemistry. Pathol Int 2017;67:398-403. [Crossref] [PubMed]
- Piessen G, Messager M, Leteurtre E, et al. Signet ring cell histology is an independent predictor of poor prognosis in gastric adenocarcinoma regardless of tumoral clinical presentation. Ann Surg 2009;250:878-87. [Crossref] [PubMed]
- Postlewait LM, Squires MH, Kooby DA, et al. The Prognostic Value of Signet-Ring Cell Histology in Resected Gastric Adenocarcinoma. Ann Surg Oncol 2015;22:S832-9. [Crossref] [PubMed]
- Hsu JT, Wang CW, Le PH, et al. Clinicopathological characteristics and outcomes in stage I-III mucinous gastric adenocarcinoma: a retrospective study at a single medical center. World J Surg Oncol 2016;14:123. [Crossref] [PubMed]
- Kunisaki C, Akiyama H, Nomura M, et al. Clinicopathologic characteristics and surgical outcomes of mucinous gastric carcinoma. Ann Surg Oncol 2006;13:836-42. [Crossref] [PubMed]
- Tsuruta S, Kohashi K, Yamada Y, et al. Solid-type poorly differentiated adenocarcinoma of the stomach: Deficiency of mismatch repair and SWI/SNF complex. Cancer Sci 2020;111:1008-19. [Crossref] [PubMed]
- Kunisaki C, Akiyama H, Nomura M, et al. Clinicopathological properties of poorly-differentiated adenocarcinoma of the stomach: comparison of solid- and non-solid-types. Anticancer Res 2006;26:639-46. [PubMed]
- Zhang S, Tong YX, Zhang XH, et al. A novel and validated nomogram to predict overall survival for gastric neuroendocrine neoplasms. J Cancer 2019;10:5944-54. [Crossref] [PubMed]
- Fang C, Wang W, Feng X, et al. Nomogram individually predicts the overall survival of patients with gastroenteropancreatic neuroendocrine neoplasms. Br J Cancer 2017;117:1544-50. [Crossref] [PubMed]
- Adachi Y, Yasuda K, Inomata M, et al. Pathology and prognosis of gastric carcinoma: well versus poorly differentiated type. Cancer 2000;89:1418-24. [Crossref] [PubMed]
- Cronin KA, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results (SEER) Program of the National Cancer Institute. Cancer 2014;120:3755-7. [Crossref] [PubMed]
- Wang Q, Liu G, Hu C. Molecular Classification of Gastric Adenocarcinoma. Gastroenterology Res 2019;12:275-82. [Crossref] [PubMed]
- In H, Solsky I, Palis B, et al. Validation of the 8th Edition of the AJCC TNM Staging System for Gastric Cancer using the National Cancer Database. Ann Surg Oncol 2017;24:3683-91.
- Shu P, Qin J, Shen K, et al. The IGCA staging system is more accurate than AJCC7 system in stratifying survival of patients with gastric cancer in stage III. BMC Cancer 2017;17:238. [Crossref] [PubMed]
- Jung H, Lee HH, Song KY, et al. Validation of the seventh edition of the American Joint Committee on Cancer TNM staging system for gastric cancer. Cancer 2011;117: 2371-2378.
- Sano T, Coit DG, Kim HH, et al. Proposal of a new stage grouping of gastric cancer for TNM classification: Inter-national Gastric Cancer Association staging project. Gastric Cancer 2017;20:217-25. [Crossref] [PubMed]
- Son T, Sun J, Choi S, et al. Multi-institutional validation of the 8th AJCC TNM staging system for gastric cancer: Analysis of survival data from high-volume Eastern centers and the SEER database. J Surg Oncol 2019;120:676-84. [Crossref] [PubMed]
- Wang Y, Li J, Xia Y, et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol 2013;31:1188-95. [Crossref] [PubMed]
- Song W, Lv CG, Miao DL, et al. Development and validation of a nomogram for predicting survival in patients with gastrointestinal stromal tumours. Eur J Surg Oncol 2018;44:1657-65. [Crossref] [PubMed]



