
MicroRNA-221 promotes tumor progression by targeting HHIP in human glioblastoma
Introduction
Glioblastoma (GBM) is the most common type of brain tumor and is a worldwide health problem (1-3). The clinical symptoms of patients with GBM include include headache, nausea and vomiting. Annually, GBM has an incidence rate of approximately two to three cases per 100,000 worldwide (2-5). Despite major advances in diagnosis and treatment, the relative total 5-year survival rate remains below 10% (6-8). Recent studies have showed that complex cellular and molecular events may be implicated in prognosis of GBM. Therefore, there is an urgent need to explore the molecular and cellular alterations associated with GBM in order to develop novel therapies.
MicroRNAs (miRNAs) are endogenous, single-stranded, non-coding, and small RNA molecule groups of 17–25 nucleotides in length (9-13). They mainly bind to the 3’-untranslated region (3’-UTR) of mRNA target genes for the regulation of gene expression. Previous research has confirmed that the function of miRNAs is linked to the regulation of pathological and physiological processes such as drug resistance, lipid metabolism, oncogenesis, apoptosis, cell differentiation, cell cycle distribution, cell growth, and cell migration and invasion (9-13). Moreover, miRNAs are found to be aberrantly expressed in many different tumor types including bladder, colorectal, gastric and cervical cancers (10-12). miR-221, a newly discovered tumor-related miRNA, is abnormally expressed in a variety of tumor types (11-13). However, the underlying molecular mechanism, biological role, and expression of miR-221 in GBM remains largely unexplored. This study primarily attempted to ascertain the expression level of miR-221 in GBM and to evaluate its role and potential mechanisms. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-21-99).
Methods
Tissue collection
Fifty-two GBM tissues were harvested from cases that had surgical procedures at our hospital between March 2012 and March 2018. Twenty normal brain tissue were harvested from the patients who underwent intracranial decompression and excision due to brain injury or cerebral hemorrhage at our hospital between March 2012 and March 2018. The tissue samples were incubated at −80 °C for subsequent experiments. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Clinical Research Ethics Committee of Harbin Medical University Cancer Hospital (No. 2019zlyy0168). Written informed consent form was obtained from all patients.
Cell culture and transfection
The Chinese Academy of Sciences Cell Bank provided the normal human astrocytes (NHA) and three GBM cell lines (A172, U251, and T98G). Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fisher Scientific, USA) with 10% fetal bovine serum (FBS) was used to culture the GBM cells which were stored in 5% CO2 at 37 °C in a humidified chamber.
A miR-221 inhibitor, negative control (NC), and small interfering RNA targeting human hedgehog-interacting protein (siHHIP) were provided by GenePharma (Shanghai, China). The U251 cells were transfected using the NC, miR-221 inhibitor, and siHHIP, or cotransfected with the miR-221 inhibitor and HHIP siRNA (miR-221 inhibitor + siHHIP) with the use of a Lipofectamine™ 2000 reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Real-time PCR
Trizol Reagent (Alphainnotech, USA) was used to extract the total RNA of cells or tissues. Real-time PCR reactions were conducted with a PrimeScript® miRNA RT-PCR Kit (Takara, Dalian, China) and a 7900HT Real-Time PCR System (Applied Biosystems) in line with the manufacturer’s protocols. U6 acted as internal references. All experiments were performed in triplicate.
Cell proliferation assay
For the MTT [3-(4,5-dimethyl-2-thiazolyl) -2,5-diphenyl-2-H-tetrazolium bromide] assay, 10,000 transfected cells/well in 96-well plates were cultured for 24, 48, and 72 hours. Subsequently, the cells in each well were fixed with 0.5 mg/mL of MTT (Sigma-Aldrich, St. Louis, MO, USA) and kept for 4 hours at 37 °C. Next, the supernatant was removed, and 150 mM of dimethyl sulfoxide (DMSO; Sigma-Aldrich, USA) was introduced. An enzyme-linked immune detector (Alphainnotech Company, USA) was used to detect the absorbance value of each well at 490 nm. All experiments were performed in triplicate.
Colony formation assay
U251 cells that were transfected with the miR-221 inhibitor, NC, or siHHIP were plated onto six-well plates. Five hundred transfected cells/well in six-well plates were cultured for two weeks. The colonies were fixed and then stained in turn with methanol and crystal violet, and the number of colonies was calculated.
Wound healing assay
A wound healing assay of U251 cells, transfected with the miR-221 inhibitor or NC, was performed according to the method described in our previous study (8). The wound closure distance (in contrast to the control at 0 hours) was ascertained.
Transwell invasion assays
The assay was conducted by a 24-well insert with 8 µm pores that were coated with Matrigel. The upper filter was seeded with transfected cells (0.2 mL) at 5×105 cells/mL density of serum-free medium. An amount of 500 µL DMEM supplemented by 10% FBS was introduced into the low chamber as the chemo-attractant. After 24 hours of incubation, the U251 cells on the upper side were washed away with PBS. The membranes were fixed with methanol and stained with eosin and hematoxylin. The number of cells that had migrated through the Transwell membrane in different groups was counted.
MicroRNA target prediction and luciferase reporter assay
TargetScan 7.1 (www.targetscan.org) was used to predict miR-221’s probable targets. We conducted the luciferase reporter assay according to the methodology described in our previous study (8). The results were the average of three duplicates and were repeated three times.
Western blot analysis
All cells underwent the lysing process directly with 1×SDS loading buffer [2% sodium dodecyl sulfate (SDS), 10% glycerol, 0.1% bromophenol blue, 100 mM dithiothreitol (DTT), pH 6.8, and 50 mM Tris-Cl as the whole-cell sample. Immunoblotting was carried out with primary antibodies (Santa Cruz Biotechnology, Dallas, TX, USA) against HHIP (1:1,000) and β-actin (1:1,000). HRP-labeled goat anti-mouse IgG (Beyotime, Shanghai, China) was used to incubate the membranes at room temperature for 2 hours. Finally, with the use of the improved chemiluminescence (ECL Plus; Amersham Pharmacia Biotech, Piscataway, NJ, USA), we viewed the bands. The results were the average of three duplicates and were repeated three times.
Xenograft tumor model
Nude mice were provided by the Beijing Laboratory Animal Center. All experimental procedures were approved by Committee of Harbin Medical University for Animal Care (No. 20190051024) and use in accordance with National Guide on Animal Care and Use. The mice were separated into two groups of five mice each. The nude mice was subcutaneously injected U251 cells (2×106) transfected with either the miR-221 inhibitor or the negative control. A caliper was used to measure the tumor volume every week according to the following equation: volume = (width2 × length)/2.
Statistical analysis
Data are denoted as the mean ± standard deviation (SD). One-way ANOVA or Student’s t-tests were used to analyze the data using the SPSS 19.0 software package (IBM SPSS, Armonk, NY, USA). A P value <0.05 was considered to be statistically significant.
Results
MiR-221 expression was upregulated in GBM cell lines and tissues
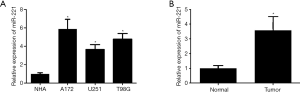
We conducted RT-PCR for the detection of miR-221 expression. There was a noticeable increase of miR-221 expression in A172, U251, and T98G cells compared to that in NHA cells (Figure 1A). Furthermore, miR-221 upregulation was also found in GBM tissues compared with that in normal brain tissue (Figure 1B). According to these results, miR-221 would appear to be vital to the progression of GBM.
MiR-221 promotes proliferation of GBM cells
To further confirm the effects exerted by miR-221 on GBM cells, U251 cells were transfected using either the miR-221 inhibitor or the NC. RT-PCR confirmed that miR-221 was downregulated in U251 cells transfected with the miR-221 inhibitor in contrast to cells transfected with the NC (Figure 2A). Furthermore, according to the results of the MTT and colony formation assays, miR-221 downregulation noticeably reduced the ability of U251 cells to proliferate (Figure 2B,C,D). These results suggest that miR-221 facilitates the proliferation of GBM cells in vitro.

miR-221 enhances GBM cell migration and invasion
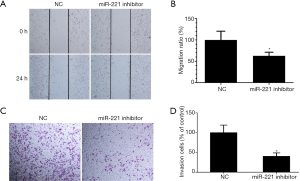
We conducted wound healing and Transwell invasion assays to determine the influence of miR-221 on the migration and invasion behavior of U251 cells. The results of the wound healing assay revealed that knockdown of miR-221 noticeably hindered the migration of U251 cells (Figure 3A,B). Similarly, knockdown of miR-221 also remarkably reduced the invasive potential of U251 cells (Figure 3C,D). Hence, the findings above suggest that miR-221 facilitates GBM cell migration and invasion.
HHIP is a direct target of miR-221 in GBM cells
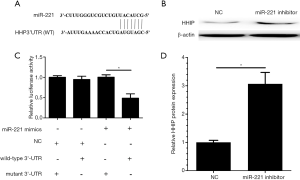
In order to identify the mechanisms of miR-221 on GBM progression, we searched the TargetScan database and identified HHIP as a probable miR-221 target gene (Figure 4A). Next, the interaction between HHIP and miR-221 was estimated by using dual luciferase reporter and western blot assays. When cotransfected with HHIP 3’-UTR luciferase reporter plasmid, miR-221 mimics caused an evident reduction in the luciferase activity of HHIP, while miR-221 mimics showed no effect on the mutated 3’-UTR fragment (Figure 4B). Meanwhile, the results of the western blot assay showed that the HHIP protein level was upregulated during the reduction of miR-221 (Figure 4C,D). According to these results, miR-221 was shown to play a promoting role in GBM cells by targeting HHIP.
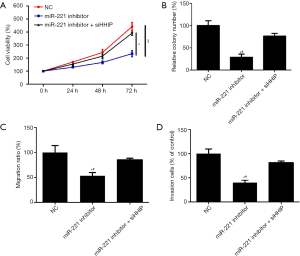
Alteration of HHIP expression influences the effects of miR-221 on GBM cells
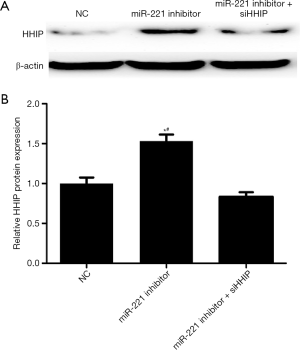
To validate our hypothesis that miR-221 influenced the behavior of GBM cells by targeting HHIP, siHHIP and miR-221 inhibitors were cotransfected to the U251 cells. Western blotting confirmed the knockdown of HHIP (Figure 5). In addition, the results of the rescue experiments showed that HHIP silencing rescued the suppressing influence exerted by the miR-221 suppressor on the proliferation, migration, and invasion behavior of GBM cells (Figure 6). Our data implied that HHIP was a downstream functional effector of miR-221.
miR-221 expression promotes GBM growth in vivo
A subcutaneous xenograft model was generated in nude mice to validate the influence exerted by miR-221 on GBM development in vivo. Compared with the control group, mice treated with a miR-221 inhibitor showed a decrease in tumor volume and weight (Figures 7A,B,C). These data suggested that a reduction of miR-221 was able to inhibit GBM growth in vivo.

Discussion
Numerous studies have identified that alteration of miRNA expression is involved in the development of several tumors, including GBM (8-14). In the current study, we found a noticeable upregulation in miR-221 expression in GBM cell lines and GBM tissues. Furthermore, decreased miR-221 expression markedly inhibited the proliferation, colony formation, migration, and invasion of GBM cells. Further investigations confirmed that miR-221 functioned by targeting HHIP and that knocking down HHIP expression could reverse the effect of a miR-221 inhibitor on GBM cell proliferation, colony formation, migration and invasion. In addition, we also found that reduction of miR-221 expression led to the suppression of GBM growth in vivo. As far as we know, this is the first report demonstrating the oncogenic role of miR-221 in GBM.
Previous studies have illustrated that miR-221 is involved in the pathological process of malignant tumors (12,14-16). For instance, Liu et al. suggested that miR-221 expression was increased in hepatocellular carcinoma tissues, and downregulation of miR-221 inhibited cell migration in vitro (14). Mohammadian et al. reported that miR-221 promoted the proliferation and radioresistance of gastric carcinoma cells (15), and Zhao et al. found that upregulation of miR-221 was correlated with lung metastasis, tumor grade, and pathological stage in patients with osteosarcoma (12). Similarly, a high level of mir-221-3p was associated with metastatic activity and malignant potential in patients with colorectal cancer (16). In this study, our results indicated that miR-221 plays an oncogenic role in GBM progression.
To date, only a few targets of miR-221 have been confirmed. For instance, Mukohyama et al. found that QKI-5, an oncogene, was a downstream functional target of miR-221 in colorectal cancer stem cells (11). Gong et al. demonstrated that miR-221 promoted apoptosis of lens epithelial cells by targeting SIRT1 and E2F3 (10). In our study, HHIP was identified as another target of miR-221 through luciferase reporter and western blot assays. HHIP, mapping to chromosome 4q31.21–31.3, negatively regulates activation of the hedgehog signaling pathway and functions as an anti-oncogene in many kinds of tumors, including gastric, colon, lung, and breast cancers (17-21). HHIP is critical to many biological functions linked to tumor development, such as epithelial-mesenchymal transition, metastasis, apoptosis and proliferation (18-22). In GBM-related studies, Shahi et al. found that HHIP expression was lower in GBM, and its lower expression was related to methylation of gene promoters (22). Our previous study also revealed that HHIP was downregulated in GBM, and abnormal HHIP expression was closely related to poor prognosis (23). In this study, we also found that knocking down HHIP expression reversed the effect of miR-221 on GBM cell invasion, migration, colony formation, and proliferation. Overall, the combination of these findings leads us to conclude that HHIP acts as a target of miR-221 and is involved in regulating the proliferation, migration, and invasion of GBM cells.
Conclusions
In conclusion, we found that miR-221 was upregulated in GBM and actively enhanced tumor progression by targeting HHIP. These findings may suggest a potential therapeutic target for GBM.
Acknowledgments
Funding: This work is supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-21-99
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-21-99
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-21-99). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by Clinical Research Ethics Committee of Harbin Medical University Cancer Hospital (No. 2019zlyy0168). Written informed consent form was obtained from all patients. All experimental procedures were approved by Committee of Harbin Medical University for Animal Care (No. 20190051024) and Use in accordance with National Guide on Animal Care and Use.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Davies J, Reyes-Rivera I, Pattipaka T, et al. Garcia J and Burton E: Survival in elderly glioblastoma patients treated with bevacizumab-based regimens in the United States. Neurooncol Pract 2018;5:251-61. [Crossref] [PubMed]
- Liang H, Chen G, Li J, et al. Snail expression contributes to temozolomide resistance in glioblastoma. Am J Transl Res 2019;11:4277-89. [PubMed]
- Yusuf MB, Gaskins J, Amsbaugh MJ, et al. Survival impact of prolonged postoperative radiation therapy for patients with glioblastoma treated with combined-modality therapy. Neurooncol Pract 2019;6:112-23. [Crossref] [PubMed]
- Davies J, Reyes-Rivera I, Pattipaka T, et al. Survival in elderly glioblastoma patients treated with bevacizumab-based regimens in the United States. Neurooncol Pract 2018;5:251-61. [Crossref] [PubMed]
- Yang L, Zhang H. Expression of Cytosolic Phospholipase A2 Alpha in Glioblastoma Is Associated With Resistance to Chemotherapy. Am J Med Sci 2018;356:391-8. [Crossref] [PubMed]
- MacLeod RAF, Schneider B, Sivakova I, et al. High level EGFR amplification in a newly established glioblastoma cell line 170-MG-BA. Neoplasma 2019;66:109-17. [Crossref] [PubMed]
- Allani SK, Weissbach H, Lopez TMA. Sulindac induces differentiation of glioblastoma stem cells making them more sensitive to oxidative stress. Neoplasma 2018;65:376-88. [Crossref] [PubMed]
- Chang L, Lei X, Qin YU, et al. MicroRNA-133b inhibits cell migration and invasion by targeting matrix metalloproteinase 14 in glioblastoma. Oncol Lett 2015;10:2781-6. [Crossref] [PubMed]
- Cao S, Lin L, Xia X, et al. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget 2017;9:321-31. [Crossref] [PubMed]
- Gong W, Li J, Wang Y, et al. miR-221 promotes lens epithelial cells apoptosis through interacting with SIRT1 and E2F3. Chem Biol Interact 2019;306:39-46. [Crossref] [PubMed]
- Mukohyama J, Isobe T, Hu Q, et al. miR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res 2019;79:5151-8. [Crossref] [PubMed]
- Zhao H, Yan P, Wang J, et al. Clinical significance of tumor miR-21, miR-221, miR-143, and miR-106a as biomarkers in patients with osteosarcoma. Int J Biol Markers 2019;34:184-93. [Crossref] [PubMed]
- Iida M, Hazama S, Tsunedomi R, et al. Overexpression of miR 221 and miR-222 in the cancer stroma is associated with malignant potential in colorectal cancer. Oncol Rep 2018;40:1621-31. [Crossref] [PubMed]
- Liu Z, Wang C, Jiao X, et al. miR-221 promotes growth and invasion of hepatocellular carcinoma cells by constitutive activation of NFκB. Am J Transl Res 2016;8:4764-77. [PubMed]
- Mohammadian F, Pilehvar-Soltanahmadi Y, Mofarrah M, et al. Down regulation of miR-18a, miR-21 and miR-221 genes in gastric cancer cell line by chrysin-loaded PLGA-PEG nanoparticles. Artif Cells Nanomed Biotechnol 2016;44:1972-8. [Crossref] [PubMed]
- Yuan K, Xie K, Fox J, et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology 2013;145:853-64.e9. [Crossref] [PubMed]
- Zhou X, Qiu W, Sathirapongsasuti JF, et al. Gene expression analysis uncovers novel hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics 2013;101:263-72. [Crossref] [PubMed]
- Zuo Y, Lv Y, Qian X, et al. Inhibition of HHIP Promoter Methylation Suppresses Human Gastric Cancer Cell Proliferation and Migration. Cell Physiol Biochem 2018;45:1840-50. [Crossref] [PubMed]
- Agrawal V, Kim DY, Kwon YG. Hhip regulates tumor-stroma-mediated upregulation of tumor angiogenesis. Exp Mol Med 2017;49:e289. [Crossref] [PubMed]
- Paluszczak J, Wiśniewska D, Kostrzewska-Poczekaj M, et al. Prognostic significance of the methylation of Wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck squamous cell carcinomas. Clin Oral Investig 2017;21:1777-88. [Crossref] [PubMed]
- Song Y, Zuo Y. Occurrence of HHIP gene CpG island methylation in gastric cancer. Oncol Lett 2014;8:2340-4. [Crossref] [PubMed]
- Shahi MH, Zazpe I, Afzal M, et al. Epigenetic regulation of human hedgehog interacting protein in glioma cell lines and primary tumor samples. Tumour Biol 2015;36:2383-91. [Crossref] [PubMed]
- Chang L, Zhang P, Zhao D, et al. The hedgehog antagonist HHIP as a favorable prognosticator in glioblastoma. Tumour Biol 2016;37:3979-86. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


