
Annexin A7 and JNK knockdown suppress the lymphatic metastasis potential of hepatocellular carcinoma cells: Possible contributions of ATF2 and sequence-related lncRNA NONMMUT114121.1
Introduction
Hepatocellular carcinoma (HCC) is one of the five most common malignant tumors globally and has the second highest mortality rate among neoplastic diseases (1). Lymph node metastasis (LNM) is a seminal event in HCC mortality, so elucidating the underlying molecular mechanisms will have a profound impact on diagnosis and treatment (2). The mouse ascites syngeneic HCC line Hca-P has proven to be a valuable model for studying LNM in vitro (3-5), and our previous study found that the calcium-dependent phospholipid binding protein Annexin A7 (ANXA7) and c-Jun NH2-terminal kinase (JNK) both contribute to in vitro LNM capacity (6,7).
Annexin A7, also termed ANXA7 or synexin, functions as a Ca2+-activated GTPase in membrane fusion (7), thereby contributing to cell motility and proliferation. ANXA7 protein is widely expressed in the liver in subtypes of 47 kDa and 51 kDa (8). Downregulation of ANXA7 decreases the proliferation, migration, invasion, apoptosis, and lymphatic adherence of HCC cells in vitro (4,9-12), strongly suggesting that ANXA7 signaling serves to drive LNM.
JNK, a member of the mitogen activated protein kinase (MAPK) family, is also a critical regulator of HCC cell death and a potential target for HCC therapy (13,14). We previously reported that JNK expression at both mRNA and protein levels was greater in a HCC cell line with high lymphatic metastasis (Hca-F >70%) compared to a HCC cell line with low lymphatic metastasis (Hca-P <30%) (6,15). JNK is activated by phosphorylation (16) and then translocates to the nucleus where it phospho-activates various transcription factors implicated in oncogenesis (17,18). In HCC cells, JNK downregulation reduced ANXA7 expression, while ANXA7 downregulation had no effect of JNK expression (our unpublished observations), suggesting that ANXA7 expression is controlled by JNK signaling.
Activating transcription factor-2 (ATF2), a member of the activating protein-1 (AP-1) transcription factor family, contains a basic leucine zipper (bZIP) domain for dimerization at the C-terminus and a basic domain for DNA binding at the N-terminus that is also a substrate for JNK phosphorylation (19,20). Binding to cis-acting elements in the form of dimers or trimers promotes expression of genes that can facilitate malignant proliferation (21). However, ATF2 has the characteristics of both an oncogene and tumor suppressor in different tissues, which may depend on its subcellular localization to the nucleus or cytoplasm. Nuclear or cytoplasmic localization is regulated by protein kinase (PKC) epsilon (22). Cell proliferation, migration, and invasion were significantly reduced and apoptotic rate significantly increased by silencing ATF2 in HCC cells (23).
Long non-coding RNAs (lncRNAs) are RNA molecules >200 nt that do not encode proteins but rather regulate expression of sequence-related mRNAs. Recent studies have identified several lncRNAs as epigenetic regulators controlling the expression of genes involved in multiple biological processes (24-26). Our recent study found that lncRNA NONMMUT114121.1 was associated with ATF2 by sequencing.
ANXA7, JNK, and ATF2 appear critical for regulation of HCC activity, but the precise signaling mechanisms remain poorly understood. To the best of our knowledge, there have been no studies examining the effect of ANXA7 and JNK expression levels on ATF2 expression and associated changes in HCC cell LNM capacity. Therefore, we assessed the effects of shRNA-mediated ANXA7 and JNK knockdown on ATF2 activity and expression of its sequence-related lncRNA NONMMUT114121.1, and subcellular distribution as well as related effects on HCC cell lymphatic metastasis capacity.
Methods
Cell lines and cell culture
The mouse HCC cell line Hca-P with low lymphatic metastatic potential (generously presented by Prof. Jianwu Tang; Department of Pathology, Dalian Medical University, Dalian, Liaoning, China) was established and maintained by the Key Laboratory of Tumor Metastasis in Liaoning Province. Hca-P cells were routinely cultured in RPMI 1640 (Invitrogen, USA) medium containing 1% gentamicin/streptomycin (Beyotime, China) and 10% fetal bovine serum (Gibco, USA), and placed in 37 °C incubator (Thermo, USA) with 5% CO2 atmosphere at 37 °C.
Transient and stable transfection
Transfection of targeted shRNA expression vectors down-regulated the expression levels of ANXA7 and JNK in Hca-P cells. Concisely, the shRNA-ANXA7 expression plasmid 5’-CACCGTCAGAATTGAGTGGGAATTTCAAGAGAATTCCCACTCAATTCTGACTTTTTTG-3’ and the shRNA-JNK expression plasmid 5’-CACCGCAGGCCTAAATACGCTGGATTCAAGAGATCCAGCGTATTTAGGCCTGTTTTTTG-3’ were inserted separately into the pGPU6/GFP/Neo-shRNA expression vector [expressing green fluorescent protein (GFP) and the Neo resistance cassette], yielding pGPU6/GFP/Neo-shRNA-ANXA7 (termed shRNA-ANXA7) and pGPU6/GFP/Neo-shRNA-JNK (shRNA-JNK), respectively. A nonspecific shRNA pGPU6/GFP/Neo vector was used as a control (control-shRNA) for both targeted vectors. Plasmids were designed by Genepharma Co., Ltd. (Shanghai, China). Hca-P cells were inoculated in 24-well plates at a rate of 5×105/well with serum-free RPMI-1640 medium overnight and then transfected with the indicated vector using Lipofectamine 2000 (Invitrogen, Cat. 11668019, USA) according to the manufacturer’s instructions. After 24 and 48 h, the transfection efficiency was examined by fluorescence microscopy (Olympus, Japan). The transfected cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (Gibco) and 400 µg/mL G418 (Gibco) for 28 days to obtain stable transfected cell lines. During the selection process, part of the medium is changed daily. When the cells approached the junction point, they were divided into two or three parts of the transfection medium and maintained as described.
Establishment and Sequencing of RNA Library
Total RNA was extracted from Hca-P cells according to the instructions using Trizol reagent (TakaRa, Japan). RNA samples were stored on dry ice and sent to Shanghai Biotechnology Corporation for database construction and sequencing using Illumina HiSeq Xten in the PE150 sequencing mode, yielding 15G of data.
Analysis of cell invasion
Cell invasion was assessed by transwell migration assays. Transwell chamber units were incubated with Matrigel Matrix (Corning, USA) at 37 °C for 1 h to generate an artificial basement membrane, and then rehydrated with 10 µL of serum-free RPMI 1640 medium. The upper chambers of the inserts were seeded with 4×104 cells stably transfected with control-shRNA, shRNA-ANXA7, or shRNA-JNK as indicated in 100 µL serum-free RPMI 1640 medium, whilst the lower chambers were filled with 500 µL of RPMI 1640 medium containing 20% FBS as a chemoattractant. After 48 h of incubation at 37 °C under a humidified 5% CO2 atmosphere, the transwell inserts were swabbed on the upper chamber side to remove non-invasive cells, fixed, stained with crystal violet, and then examined under an inverted light microscope (Leica, Japan) to assess cell numbers on the lower chamber side. Cell numbers in 5 randomly chosen high-power fields were counted per insert as an index of cell migration.
Lymph adherence assay
Lymph nodes of the inguinal, axillary, or popliteal fossa were obtained from clean grade 615 mice and prepared as frozen sections. Sections were air dried at room temperature and placed on slides in a pre-sterilized wet box with sterile phosphate-buffered saline (PBS). Lymph node sections were then seeded with the indicated Hca-P transfection group (control-shRNA, shRNA-ANXA7, shRNA-JNK) at 2×105 cells per section and incubated at 37 °C for 24 h under 5% CO2. Seeded lymph node sections were then washed three times with pre-cooled PBS (3 min/wash), stained with HE, sealed in neutral resin, and examined under an optical microscope (Leica, Japan) to assess the density of adherent cells.
RNA extraction and gene expression analysis
The stably transfected cells were rinsed twice with ice-cold PBS. According to the manufacturer's instructions, the total RNA was extracted by using the Total RNA Extraction Kit Fast200 (Feijie, China). RNA concentration was determined by the absorbance at 260 nm. The 500-ng mRNA samples from each group were reverse-transcribed with an All-in-one First strand cDNA Synthesis Kit (Transgen, China) at 42 °C for 15 minutes and 85 °C for 5 seconds. The expression of ATF2 was detected by quantitative reverse transcription–polymerase chain reaction (qRT−PCR) using the Power SYBR Green PCR Master Mix (Thermo Fisher, USA). Relative expression was calculated by comparing ∆∆Ct method and GAPDH as the internal control. The primer sequences for ATF2, lncRNA NONMMUT114121.1, and GAPDH are listed in Table 1.
Table 1
| Gene | Forward primer | Reverse primer |
|---|---|---|
| ATF2 | 5’-AAGCTGCTTTGACCCAGCAA-3’ | 5’-GAGAAGCCGGAGTTTCTGTAGTG-3’ |
| NONMMUT114121.1 | 5’-GCGTTAGCAGTCCAGAGTCCAT -3’ | 5’-GTTTCCTCTACATCGGTTAACTTCGA -3’ |
| GAPDH | 5’-AGGTCGGTGTGAACGGATTTG-3’ | 5’-TGTAGACCATGTAGTTGAGGTCA-3’ |
Western blotting
Cells from control-shRNA, shRNA-ANXA7, and shRNA-JNK transfection groups were collected and lysed in RIPA buffer (Beyotime, China) containing phenylmethanesulfonyl fluoride (PMSF, Beyotime, China). Total protein content was measured using a bicinchoninic acid (BCA) protein assay kit (Beyotime, China). Briefly, absorbance values were measured on a microplate spectrophotometer (Thermo, USA) and compared to a standard curve. Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 10% gels and electrotransferred to polyvinylidene difluoride (PVDF) membranes (Invitrogen, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline supplemented with 0.5% Tween-20 (TBS-T) at room temperature for 1 h. Blocked membranes were then incubated at 4 °C overnight with a polyclonal antibody against ATF2 (1:600; Proteintech Cat# 11908-1-AP, RRID:AB_10695892, USA) and then with a monoclonal antibody against GAPDH (1:2000; Proteintech Cat# 60004-1-Ig, RRID:AB_2107436, USA) as the gel loading control. Membranes were washed three times in TBS-T (10 min/wash) and then incubated sequentially for 1 h each at 37 °C with goat anti-rabbit and anti-mouse secondary antibodies (both 1:20000; ZSGB-Bio Cat# ZB-2301, RRID:AB_2747412 and ZSGB-Bio Cat# ZB-2305, RRID:AB_2747415, China). Protein bands were visualized using electrochemiluminescence (ECL) reagent (Beyotime, China) and quantified by a chemiluminescence detection system.
Immunofluorescence
Stably transfected cells were washed twice with pre-cooled PBS, seeded on poly-L-lysine-coated glass slides, fixed in 10% formalin, permeabilized using 0.2% Triton-X100, blocked in 5% bovine serum albumin for 1 h, and incubated with anti-ATF2 antibody (1:100, Proteintech Cat# 11908-1-AP, RRID:AB_10695892, USA) overnight at 4 °C. The following day, cells were incubated with anti-rabbit IgG (H+L) secondary antibody (1:100, Thermo Fisher Scientific Cat# 31670, RRID:AB_228342, USA) for 1 h at room temperature. Finally, the cells were counterstained with 5 µg/mL DAPI (Beyotime, China) for 20 min and examined under an inverted epifluorescence microscope (Olympus, Japan).
Statistical analysis
All statistical analyses were performed using SPSS software (version 22.0, IBM, USA). All data are expressed as the mean ± standard deviation (SD). Independent sample t-tests were used for grouping mean comparison. A P<0.05 (two-tailed) was considered statistically significant for all tests.
Results
Determination of transfection efficiency
Transfection efficiency was evaluated by GFP emission from the pGPU6/GFP/Neo expression vector. Examination under an inverted fluorescence microscope (Olympus, Japan) revealed that after 48 h of transfection about 70% of the survival cells in all three treatment groups (control-shRNA, shRNA-ANXA7 and shRNA-JNK) emitted green fluorescence (Figure 1A). ShRNA-mediated stable transfected cell lines with ANXA7 or JNK knockdown were established by G418 screening for 28 consecutive days to examine the effects on ATF2 expression, metastasis capacity, and lymphatic adherence.
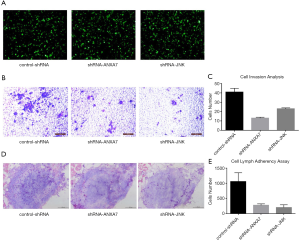
Downregulation of ANXA7 and JNK inhibited Hca-P cell invasive potential
Transwell migration assays using Matrigel as an artificial extracellular matrix revealed significantly reduced invasion potential in both ANXA7 and JNK knockdown lines compared to the control-shRNA line as indicated by crystal violet staining and cell counting (13±0.816 and 23±0.816 cells/field vs. 41±3.266 cells/field; both P<0.05) (Figure 1B,C)
Downregulation of ANXA7 and JNK expression reduced Hca-P cell lymphatic tissue adherence
Lymphatic tissue adherence was also significantly reduced by ANXA7 and JNK knockdown compared to controls (277±38 and 205±73 cells/field vs. 1056±239 cells/field; both P<0.05) (Figure 1D,E).
Downregulation of ANXA7 and JNK reduced ATF2 mRNA and protein expression
QRT-PCR confirmed that the expression of ATF2 gene was significantly decreased after ANXA7 and JNK knockdown in Hca-P cells (32.3%±1.3% and 44.8%±7.7%, respectively of control-shRNA; both P<0.05) (Figure 2A). Similarly, western blotting revealed parallel reductions in ATF2 protein expression (control-shRNA: 0.066±0.014; shRNA-ANXA7: 0.027±0.002; shRNA-JNK: 0.030±0.007; both P<0.05 vs. control-shRNA) (Figure 2B,C).
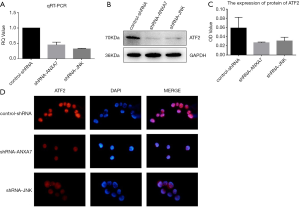
Localization of the ATF2 protein
Immunofluorescence (Figure 2D) indicated that ATF2 was mainly localized to the nucleus with substantially lower expression in the cytoplasm, while expression levels in both compartments were reduced by knockdown of ANXA7 or JNK.
Differentially expressed lncRNAs
We identified 436 unique lncRNAs differentially expressed between shRNA-JNK and control-shRNA cells, of which 236 were expressed at lower levels and 200 at higher levels in shRNA-JNK cells (Figure 3A). There were also 216 differentially expressed lncRNAs between shRNA-ANXA7 and control-shRNA cells, of which 101 were expressed at lower levels and 115 at higher levels in shRNA-ANXA7 cells (Figure 3B). The lncRNA NONMMUT114121.1 was related to ATF2 by sequence and differentially expressed in shRNA-ANXA7 cells, while expression did not differ between shRNA-JNK and control-shRNA cells.
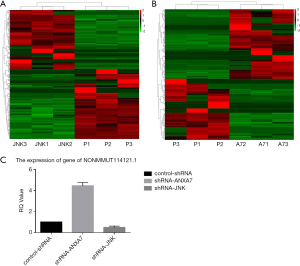
Downregulation of ANXA7 increased NONMMUT114121.1 expression
Expression of NONMMUT114121.1 was significantly increased by ANXA7 knockdown and reduced by JNK knockdown in Hca-P cells as evidenced by qRT-PCR (444.1%±32.3% and 50.0%±11.8%, respectively, of control-shRNA; both P<0.05) (Figure 3C).
Discussion
HCC is a malignant tumor with high metastatic rate. LNM is one of the most frequent metastatic patterns and markedly increases mortality risk. Tumor development, progression, and LNM are strongly associated with aberrant changes in the expression of MAPK signaling factors. ANXA7 mediates Ca2+/GTP-dependent membrane fusion, thereby contributing to secretion, cell division, growth, and apoptosis (11,27,28). Moreover, downregulation of ANXA7 strongly induced HCC cell apoptosis (29). Thus, high ANXA7 expression is a potential indicator of lymphatic metastasis risk and poor prognosis in patients with HCC as well as in gastric carcinoma patients (30). In contrast, ANXA7 acts as a tumor suppressor gene in prostate cancer and glioblastoma (31). Our previous in vitro study suggested that ANXA7 regulates the biological activities of mouse HCC cells via JNK-ANXA7 signal transduction (6). ATF2 regulates gene expression, transactivation, and nuclear export through homo-dimerization or hetero-dimerization with other AP1 family members implicated in disease pathogenesis (32,33). In this study, the invasion and lymphoid adhesion capacities of Hca-P cells were significantly reduced by knockdown of ANXA7 or JNK, and knockdown of either molecule reduced ATF2 expression at both gene and protein levels. ANXA7 activates PKC via Ca2+/GTP signaling. In turn, PKC promotes the oncogenic functions of ATF2 in the nucleus while blocking its pro-apoptotic functions in the mitochondria. Based on these findings, we speculate that ANXA7 knockdown suppresses Ca2+ and PKC signaling, which in turn reduces ATF2 activity and ensuing upregulation of genes promoting the invasive capacity and lymphatic tissue adherence of HCC cells.
JNK is a mitogen-activated protein kinase (MAPK) strongly expressed as two isoforms in liver (14). Studies in mice suggested that JNK1 initiates HCC by inducing hepatocyte apoptosis and compensatory proliferation (5,34). Diverse stimuli (such as cytokines, toxins, and drugs) activate JNK and trigger multiple downstream responses through activation of AP-1 (35). Invasion capacity is the most important characteristic determining risk of tumor-related death. In this study, we confirmed that JNK knockdown reduces the invasive and lymphatic adhesion capacities of HCC cells in vitro and concomitantly downregulates ATF2 expression at both the gene and protein levels. ATF2 is a major downstream target of JNK, so reducing JNK expression will reduce ATF2 activation via phosphorylation (16,17). ATF2 is implicated in cell cycle control, cytokine expression, and cell death. Thus, ATF2 expression and/or nuclear localization are potential markers of HCC prognosis, including LNM risk. Further, disruption of JNK-ATF2 signaling may have therapeutic efficacy against LNM HCC.
In the past 10 years, high-throughput sequencing has been widely used for lncRNA identification and analysis of specific functions in various diseases. In the current study, we focused on identifying differentially expressed lncRNAs under ANXA7 and JNK knockdown in HCC cells by sequencing to reveal novel mechanisms underlying LNM regulation. This screen identified a new lncRNA (NONMMUT114121.1) that may contribute to LNM of HCC. As shown by qRT-PCR, lncRNA NONMMUT114121.1 gene expression was significantly increased in ANXA7 knockdown Hca-P cells compared to control-shRNA Hca-P cells, suggesting that ANXA7 knockdown may affect ATF2 expression by altering lncRNA NONMMUT114121.1 expression. Thus, ATF2 modulation by lncRNA NONMMUT114121.1 may be a valuable therapeutic target worthy of further study.
In addition to expression level, targeting of proteins to specific subcellular compartments and structures is critical for biological activity. Therefore, a change in intracellular distribution may have a greater influence on activity. For instance, subcellular location may explain the heterogeneous effects of various proteins in different cell types and cancer stages. Immunofluorescence revealed that ATF2 was mainly localized to the nucleus, and nuclear expression was markedly reduced by ANXA7 or JNK knockdown. Like ANXA7, ATF2 has been shown to promote or inhibiting tumors in different tissues. For instance, ATF2 plays a carcinogenic role in melanoma (33) and prostate cancer (36) but acts as a cancer suppressor in leukemia, benign skin cancer, and breast cancer (37,38). JNK is activated by phosphorylation and then enters the nucleus, where it can bind to the N-terminal region of ATF2 and trigger transcriptional activity. Nuclear expression of ATF2 was reduced at both gene and protein levels by JNK or ANXA7 knockdown. Based on previous reports and our own experimental results, we speculate that ATF2 promotes HCC by driving the expression of proteins involved in cell migration and adhesion to lymphatic tissue.
In our previous study, GeneGo analysis predicted that ANXA7 may influence HCC through JNK signaling (6). In this study, we confirmed that knockdown of JNK or ANXA7 reduces HCC cell invasive capacity and lymphatic tissue adherence concomitant with a reduction in ATF2 expression and nuclear localization. Based on these results, we suggest that the modulation of ATF2 nuclear activity by ANXA7 and JNK is critical a regulator of hepatic carcinogenesis. These pathways are thus promising targets for therapeutic intervention.
Acknowledgments
We thank Prof. Jianwu Tang for providing us the Hca-P cell line.
Funding: This work was supported in part by grants from
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2111
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2111
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2111). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Krivtsova O, Makarova A, Lazarevich N. Aberrant expression of alternative isoforms of transcription factors in hepatocellular carcinoma. World J Hepatol 2018;10:645-61. [Crossref] [PubMed]
- Kim DW, Talati C, Kim R. Hepatocellular carcinoma (HCC): beyond sorafenib-chemotherapy. J Gastrointest Oncol 2017;8:256-65. [Crossref] [PubMed]
- Liu S, Sun MZ, Tang JW, et al. High-performance liquid chromatography/nano-electrospray ionization tandem mass spectrometry, two-dimensional difference in-gel electrophoresis and gene microarray identification of lymphatic metastasis-associated biomarkers. Rapid Commun Mass Spectrom 2008;22:3172-8. [Crossref] [PubMed]
- Jin Y, Wang S, Chen W, et al. Annexin A7 suppresses lymph node metastasis of hepatocarcinoma cells in a mouse model. BMC Cancer 2013;13:522. [Crossref] [PubMed]
- Song L, Mao J, Zhang J, et al. Annexin A7 and its binding protein galectin-3 influence mouse hepatocellular carcinoma cell line in vitro. Biomed Pharmacother 2014;68:377-84. [Crossref] [PubMed]
- Jin Y, Mao J, Wang H, et al. Enhanced tumorigenesis and lymphatic metastasis of CD133+ hepatocarcinoma ascites syngeneic cell lines mediated by JNK signaling pathway in vitro and in vivo. Biomed Pharmacother 2013;67:337-45. [Crossref] [PubMed]
- Jin YL, Wang ZQ, Qu H, et al. Annexin A7 gene is an important factor in the lymphatic metastasis of tumors. Biomed Pharmacother 2013;67:251-9. [Crossref] [PubMed]
- Ibrahim MM, Sun M-Z, Huang Y, et al. Down-regulation of ANXA7 decreases metastatic potential of human hepatocellular carcinoma cells in vitro. Biomed Pharmacother 2013;67:285-91. [Crossref] [PubMed]
- Zhao Y, Yang Q, Wang X, et al. AnnexinA7 down-regulation might suppress the proliferation and metastasis of human hepatocellular carcinoma cells via MAPK/ ERK pathway. Cancer Biomark 2018;23:527-37. [Crossref] [PubMed]
- Huang Y, Wang Q, Du Y, et al. Inhibition of annexin A7 gene and protein induces the apotosis and decreases the invasion, migration of the hepatocarcinoma cell line. Biomed Pharmacother 2014;68:819-24. [Crossref] [PubMed]
- Bai L, Guo Y, Du Y, et al. 47 kDa isoform of Annexin A7 affecting the apoptosis of mouse hepatocarcinoma cells line. Biomed Pharmacother 2016;83:1127-31. [Crossref] [PubMed]
- Wang XY, Gao F, Sun YR, et al. In vivo and in vitro effect of hepatocarcinoma lymph node metastasis by upregulation of Annexin A7 and relevant mechanisms. Tumour Biol 2016;37:911-24. [Crossref] [PubMed]
- Kuo LM, Chen PJ, Sung PJ, et al. The Bioactive Extract of Pinnigorgia sp. Induces Apoptosis of Hepatic Stellate Cells via ROS-ERK/JNK-Caspase-3 Signaling. Mar Drugs 2018;16:19. [Crossref] [PubMed]
- Li Y, Zuo H, Wang H, et al. Decrease of MLK4 prevents hepatocellular carcinoma (HCC) through reducing metastasis and inducing apoptosis regulated by ROS/MAPKs signaling. Biomed Pharmacother 2019;116:108749. [Crossref] [PubMed]
- Qazi AS, Sun M, Huang Y, et al. Subcellular proteomics: determination of specific location and expression levels of lymphatic metastasis associated proteins in hepatocellular carcinoma by subcellular fractionation. Biomed Pharmacother 2011;65:407-16. [Crossref] [PubMed]
- Kou Y, Yan X, Liu Q, et al. HBV upregulates AP-1 complex subunit mu-1 expression via the JNK pathway to promote proliferation of liver cancer cells. Oncol Lett 2019;18:456-64. [Crossref] [PubMed]
- Gozdecka M, Lyons S, Kondo S, et al. JNK suppresses tumor formation via a gene-expression program mediated by ATF2. Cell Rep 2014;9:1361-74. [Crossref] [PubMed]
- Zeke A, Misheva M, Remenyi A, et al. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiol Mol Biol Rev 2016;80:793-835. [Crossref] [PubMed]
- Watson G, Ronai ZA, Lau E. ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease. Pharmacol Res 2017;119:347-57. [Crossref] [PubMed]
- Chen J, Solomides C, Simpkins F, et al. The role of Nrf2 and ATF2 in resistance to platinum-based chemotherapy. Cancer Chemother Pharmacol 2017;79:369-80. [Crossref] [PubMed]
- Tang J, Liao Y, He S, et al. Autocrine parathyroid hormone-like hormone promotes intrahepatic cholangiocarcinoma cell proliferation via increased ERK/JNK-ATF2-cyclinD1 signaling. J Transl Med 2017;15:238. [Crossref] [PubMed]
- Lau E, Kluger H, Varsano T, et al. PKCepsilon promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Cell 2012;148:543-55. [Crossref] [PubMed]
- Luo L, Cai L, Luo L, et al. Silencing activating transcription factor 2 promotes the anticancer activity of sorafenib in hepatocellular carcinoma cells. Mol Med Rep 2018;17:8053-60. [Crossref] [PubMed]
- Shang W, Adzika GK, Li Y, et al. Molecular mechanisms of circular RNAs, transforming growth factor-beta, and long noncoding RNAs in hepatocellular carcinoma. Cancer Med 2019;8:6684-99. [Crossref] [PubMed]
- Shi H, Xu Y, Yi X, et al. Current Research Progress on Long Noncoding RNAs Associated with Hepatocellular Carcinoma. Anal Cell Pathol (Amst) 2019;2019:1534607. [Crossref] [PubMed]
- Sun L, Wang L, Chen T, et al. microRNA-1914, which is regulated by lncRNA DUXAP10, inhibits cell proliferation by targeting the GPR39-mediated PI3K/AKT/mTOR pathway in HCC. J Cell Mol Med 2019;23:8292-304. [Crossref] [PubMed]
- Ye W, Li Y, Fan L, et al. Annexin A7 expression is downregulated in late-stage gastric cancer and is negatively correlated with the differentiation grade and apoptosis rate. Oncol Lett 2018;15:9836-44. [Crossref] [PubMed]
- Yuan HF, Li Y, Ye WH, et al. Downregulation of annexin A7 decreases proliferation, migration, and invasion of gastric cancer cells by reducing matrix metalloproteinase 1 and 9 expression. Am J Transl Res 2019;11:2754-64. [PubMed]
- Huang Y, Du Y, Zhang X, et al. Down-regulated expression of Annexin A7 induces apoptosis in mouse hepatocarcinoma cell line by the intrinsic mitochondrial pathway. Biomed Pharmacother 2015;70:146-50. [Crossref] [PubMed]
- Ye W, Li Y, Fan L, et al. Effect of annexin A7 suppression on the apoptosis of gastric cancer cells. Mol Cell Biochem 2017;429:33-43. [Crossref] [PubMed]
- Leighton X, Bera A, Eidelman O, et al. Tissue microarray analysis delineate potential prognostic role of Annexin A7 in prostate cancer progression. PLoS One 2018;13:e0205837. [Crossref] [PubMed]
- Gozdecka M, Breitwieser W. The roles of ATF2 (activating transcription factor 2) in tumorigenesis. Biochem Soc Trans 2012;40:230-4. [Crossref] [PubMed]
- Lau E, Feng Y, Claps G, et al. The transcription factor ATF2 promotes melanoma metastasis by suppressing protein fucosylation. Sci Signal 2015;8:ra124. [Crossref] [PubMed]
- Das M, Garlick DS, Greiner DL, et al. The role of JNK in the development of hepatocellular carcinoma. Genes Dev 2011;25:634-45. [Crossref] [PubMed]
- Seki E, Brenner DA, Karin M. A liver full of JNK: signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 2012;143:307-20. [Crossref] [PubMed]
- Ma J, Chang K, Peng J, et al. SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J Exp Clin Cancer Res 2018;37:145. [Crossref] [PubMed]
- Bhoumik A, Fichtman B, Derossi C, et al. Suppressor role of activating transcription factor 2 (ATF2) in skin cancer. Proc Natl Acad Sci U S A 2008;105:1674-9. [Crossref] [PubMed]
- Vlahopoulos SA, Logotheti S, Mikas D, et al. The role of ATF-2 in oncogenesis. Bioessays 2008;30:314-27. [Crossref] [PubMed]


