
Low expression of AQP9 and its value in hepatocellular carcinoma
Introduction
For decades, liver cancer has been one of the most frequent cancers in the world, with a high mortality rate in patients. Unfortunately, China has more cases and a higher cancer mortality rate compared to developed countries (1,2). In general, liver cancer can be divided into three types: hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, and some rare types. The main risk factors for HCC are chronic infection with hepatitis B virus or hepatitis C virus, and some other factors such as smoking, alcoholism, obesity, and type 2 diabetes (3). HCC remains difficult to treat, and surgery is the main treatment in the clinic, although fluorouracil, sorafenib and other chemotherapy drugs are also widely used. Due to the development of surgical approaches and various targeted drugs, the mortality of patients diagnosed in the early stages of the disease has decreased; however, the overall survival (OS) of advanced HCC patients requires improvement. Due to the lack of early diagnostic biomarkers with high specificity and sensitivity, patients are often diagnosed with advanced liver cancer and cannot receive effective treatment and the prognosis of such patients is poor. In addition, the frequency of tumor metastasis is high, and eventually, these patients decide to stop treatment (4). To improve the prognosis of HCC patients, it is important to explore new HCC markers, which could also help to clarify the molecular mechanisms underlying carcinogenesis and cancer progression.
Agre first discovered aquaporins (AQPs) in 1992 (5), and since then, AQPs has been widely researched. The AQP family of water channels proteins comprise 13 members (AQP0–AQP12), which can be divided into three subfamilies: classical AQPs (water-selective: AQP0, AQP1, AQP2, AQP4, AQP5, AQP6 and AQP8) and aquaglyceroporins (glycerol channel, Glps: AQP3, AQP7, AQP9 and AQP10), and AQP11 and AQP12 (6). Several aquaglyceroporin AQPs play notable roles in cancer progression. Helicobacter pylori infection upregulates AQP3 in the stomach, and regulates the ROS–HIF-1α–AQP3–ROS loop to promote gastric cancer (7). AQP7 is a critical regulator of metabolic and signaling responses to environmental cellular stresses in breast cancer (8). AQP10 has been associated with improved OS and has been identified as a potential therapeutic target in ovarian cancer (9). Non-alcoholic fatty liver disease (NAFLD) has become increasingly common in China, but has not attracted enough attention (10). Liver steatosis is characterized by ectopic accumulation of fat and altered glycerol uptake, and interestingly, AQP9 are dysregulated in liver steatosis mice (11-13). In humans, there is evidence to suggest that AQP9 inhibits the progression of HCC via the Wnt/β-catenin pathway (14), as well as epithelial-to-mesenchymal transition (15) and the ROS-HIF-1α signaling pathway (16,17). However, the value of AQP9 in the diagnosis and prognosis of HCC as well as the changes in genes and pathways related to AQP9, immune infiltration levels and the reason for the decrease of AQP9 expression in HCC remain to be elucidated.
To reveal the clinical significance of AQP9 in HCC, we comprehensively assessed AQP9 expression and its prognostic value in patients using online databases. Moreover, we analyzed AQP9 expression in liver cancer cells and tissues and compared the results with the online databases. LinkedOmics and GSEA were used to analyze the functional network of AQP9. In addition, we predicted the candidate microRNAs that bind to AQP9. Finally, we used the Tumor Immune Estimation Resource (TIMER) to clarify the relationship between AQP9 and immune infiltration in HCC. Thus, compared with published studies, we are the first to analyze the prognostic value, and biological networks of AQP9 as well as the immune cell infiltration and possible causes of the downregulation of AQP9 in HCC.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-3158).
Methods
Oncomine database
The Oncomine database (http://www.oncomine.com) is used for translational bioinformatics studies. The website provides chip data from different tumors, and processes these data with a standardized analysis process. In this study, we obtained the data for AQP9 in different types of cancer patients from the Oncomine database. The Roessler Liver, Roessler Liver 2, and Chen Liver datasets were used in the analysis.
TIMER
TIMER (https://cistrome.shinyapps.io/timer) is used for analysis of tumor-infiltrating immune cells. Six immune cell infiltrates [B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs)] can be evaluated using this resource. We used online module in TIMER to reveal the correlations between AQP9 and different marker genes of infiltrating immune cells in HCC.
The Human Protein Atlas
The Human Pathology Atlas (https://www.prote inatlas.org) contains immunohistochemistry (IHC) data based on 44 different types of normal tissues (18). The Human Protein Atlas was searched for AQP9 expression data in different human tissues.
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu) contains clinical data from patients with 33 different cancers (19). Here, we used the UALCAN database to analyze the association of AQP9 with different clinical characteristics in HCC patients.
Kaplan-Meier Plotter database analysis
The Kaplan-Meier plotter (http://kmplot.com/analysis/) database includes 5,143 breast cancer samples, 1,816 ovarian cancer samples, 2,437 lung cancer samples, and 1,065 gastric cancer samples. This database can be used to assess the association of genes with survival using different cancer samples (20). We used the Kaplan-Meier plotter to evaluate the correlation between AQP9 and the survival of HCC patients.
c-BioPortal database analysis
c-BioPortal (https://www.cbioportal.org/) includes the cancer genomic databases from The Cancer Genome Atlas (TCGA), the International Cancer Genome Consortium (ICGC) and the Gene Expression Omnibus (GEO) (21). This database provides the clinical data of patients (22). The clinical data from TCGA were downloaded from c-BioPortal and analyzed.
Gene set enrichment analysis (GSEA)
GSEA is a computational method that determines whether a pre-defined set of genes has statistically significant, concordant differences between two biological states (23,24). TCGA gene expression data of 374 liver cancer samples were downloaded from UCSC Xena, and 55,266 genes were analyzed by GSEA to identify biological processes enriched by AQP9.
LinkedOmics
LinkedOmics (http://linkedomics.org/login.php) is a publicly available online database, which includes data from all 32 cancer type in TGCA (25). The website has three analytical modules: LinkFinder, LinkInterpreter and LinkCompare. To obtain biological insights, we used the LinkInterpreter module to perform enrichment analysis based on gene ontology, biological pathways, and network modules.
Prediction of the miRNAs that target AQP9
We used the StarBase, TargetScan, miRDB and mirDIP databases to predict interactions between miRNAs and AQP9. The shared miRNAs predicted by the four databases were used for further research.
Real-time quantitative PCR and Western blot analyses
TRIzol® reagent (Invitrogen Life Technologies, USA) was used to extract total RNA from HCC cell lines (Huh7, SNU182, Li-7and Hep3B) and the normal liver cell line QSG-7701. RevertAid First Strand cDNA Synthesis Kit (TaKaRa) was used for reverse transcription of the RNA to generate cDNA. The sequences of the primers used for RT-qPCR analysis in this study are listed in Table 1. Primers were diluted in ddH2O and real-time qPCR was performed with SYBR Master Mixture (TaKaRa) on the LightCycler 480 II (Roche) according to standard protocols. Transcriptional expression was determined as the fold change in AQP9 expression relative to β-actin (IgG, 1:1,000; AF0003, Beyotime, China). AQP9 expression levels were determined using the 2−ΔΔCq method. Western blot analysis was used to determine AQP9 protein expression in HCC cell lines and a normal human liver cell. Anti-human AQP9 (1:1,000; sc-74409, Santa Cruz Biotechnology) was used as the primary antibody. Anti-mouse immunoglobulin G (IgG, 1:2,000; A0216, Beyotime, China) was used as the secondary antibody. β‐actin was used as the loading control.
Table 1
| Gene | Primers |
|---|---|
| AQP9-F | 5'-TTGCCCAAGCTATTCTCAGTCGA-3' |
| AQP9-R | 5'-CAGAGACACCGCCAGCCACAT-3' |
| Mir-23a-3p-F | 5'-CCAATTGCGCCTTCAGGCTA-3' |
| Mir-23a-3p-R | 5'-CGGCAGAGTCCTTACCCACA-3' |
| Mir-330-3p-F | 5'-GCAGAGATTCCGTTGTCGT-3' |
| Mir-330-3p-R | 5'-GCGAGCACAGAATTAATACGAC-3' |
| β-actin-F | 5'-GGCGGCACCACCATGTACCCT-3' |
| β-actin-R | 5'-AGGGGCCGGACTCGTCATACT-3' |
Patients and cell culture
HCC and adjacent normal tissues were acquired from 45 patients, 33 males and 12 females, who had not received therapy prior to surgery during the period from 2018 to 2020 at the affiliated hospital of Nantong University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from the patients, and this research was approved by the Ethics Committee of Affiliated Hospital of Nantong University (ethical code: 2018-L006). Samples were frozen in liquid nitrogen immediately and stored at –80 °C for future use. Patients provided written informed consent prior to surgery. HCC cell lines (Hep3B, Huh-7, Li-7, and SUN182) were purchased from Shanghai Institute of Cell Biology (Shanghai, China) and a normal liver cell line (QSG-7701) was purchased from Beyotime (Shanghai, China). Cells were cultured at 37 °C under 5% CO2 in RPMI-1640 or DMEM (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific), streptomycin (100 µg/mL) and penicillin (100 U/mL).
Immunohistochemistry
In this study, a two-step staining method was used to detected AQP9 protein expression in three pairs of HCC tissues and non-carcinoma tissues. Tumor sections (4 µm thick) were incubated overnight at 4 °C with mouse anti-AQP9 (1:1,000, sc74409; Santa Cruz). Sections were then washed in phosphate-buffered saline (PBS)/0.1% Triton-X, and incubated with an HRP-labeled Goat Anti-Mouse IgG antibody (IgG, 1:50; A0216, Beyotime, China). Sections were also stained with hematoxylin after diaminobenzidine staining. The sections were then observed under an optical microscope.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 and GraphPad Prism 8.02 software. Data were represented as the mean ± standard deviation (SD) and differences between groups of data were evaluated by one-way analysis of variance (ANOVA). All experiments were performed in triplicate.
Results
AQP9 expression levels in different types of human cancers
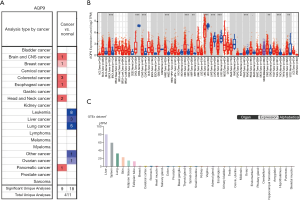
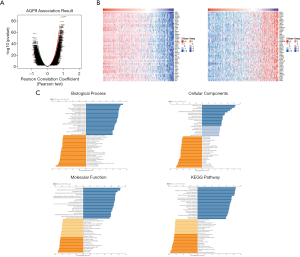
AQP9 expression levels in different tumors and corresponding normal tissues were revealed by searching the Oncomine database. In cancers of the brain and central nervous system cancer, breast cancer, esophageal cancer, head and neck cancer, colorectal cancer, and pancreatic cancer, AQP9 expression was higher than that in the corresponding normal tissues. In leukemia, liver cancer, lung cancer, and ovarian cancer, AQP9 was expressed at lower levels than that in the corresponding normal tissues (Figure 1A). These results suggested dysregulated expression of AQP9 in some cancers. We used TIMER to visualize the expression of AQP9 in various cancers. As shown in Figure 1B, lower AQP9 expression was seen in LIHC, CHOL, LUAD, LUSC, PRAD. Interestingly, according to The Human Pathology Atlas database, we found the highest AQP9 expression in the liver relative to other human tissues (Figure 1C). In conclusion, we found that AQP9 expression was the highest in the liver under normal conditions, but decreased in liver cancer.
AQP9 expression in HCC patients and cells
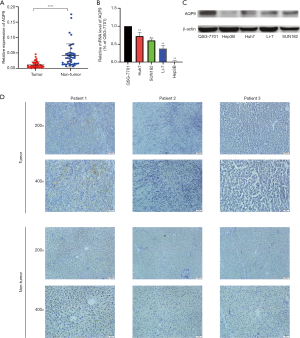
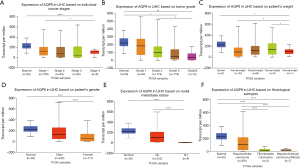
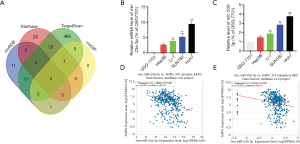
Data from the Roessler Liver, Roessler Liver 2, and Chen Liver datasets showed decreased expression of AQP9 in HCC (Figure 2). To confirm these results, we collected 45 paired liver cancer samples and adjacent non-cancerous tissues from patients during surgery. We then performed real-time-qPCR analysis of AQP9 mRNA expression in these samples. Compared with the adjacent non-tumor tissues, we detected a significant downregulation of AQP9 expression in liver cancer tissues (Figure 3A). Furthermore, we analyzed the AQP9 mRNA levels in HCC cell lines Huh7, SNU182, Li-7 and Hep3B as well as the normal human liver cell line QSG-7701 (Figure 3B,C). IHC staining revealed lower levels of AQP9 expression in tumor tissues compared with those in the adjacent non-tumor tissues (Figure 3D). As shown in Figure 4, subgroup analysis showed that AQP9 levels were significantly lower in some subgroups, such as stage, sex, weight, tumor grade, nodal metastasis status, and histological subtypes. These results indicated that AQP9 is frequently downregulated during HCC progression.

Prognostic potential of AQP9 in HCC
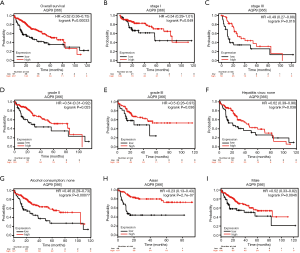
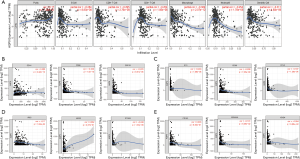
As shown in Table 2, analysis of the clinical data from TCGA showed significant differences in the expression of AQP9 in relation to AJCC tumor stage, age, sex, and alpha-fetoprotein (AFP) levels. However, differences in AQP9 expression were not significantly associated with liver fibrosis, history of HCC risk factors, Child-Pugh classification grade, cancer diagnosis first degree relative number, personal neoplasm status, and vascular invasion (P>0.05). We then explored the relationship between the AQP9 expression and clinical characteristics using the Kaplan-Meier plotter database. AQP9 expression was associated with OS in HCC patients (Figure 5A). Furthermore, low AQP9 mRNA levels were linked to worse OS in stage I, stage III, grade II, and grade III of liver cancer patients (Figure 5B-E). Low AQP9 expression was also associated with poor OS in Asian male HCC patients without alcohol consumption and with hepatitis virus infection (Figure 5F-I). Thus, considering the current incidence of liver cancer caused by NALFD in China, it can be speculated that AQP9 may impact the prognosis in these patients.
Table 2
| Parameters | Group | High groups | Low groups | P |
|---|---|---|---|---|
| Age (years) | ≤60 (n=177) | 74 | 103 | 0.003 |
| >60 (n=205) | 112 | 83 | ||
| Gender | Male (n=253) | 145 | 108 | <0.001 |
| Female (n=120) | 41 | 79 | ||
| AJCC tumor stage | T1 (n=183) | 102 | 81 | 0.018 |
| T2 (n=95) | 46 | 49 | ||
| T3 (n=80) | 33 | 47 | ||
| T4 (n=13) | 5 | 8 | ||
| AFP (ìg/L) | ≤20 (n=148) | 99 | 49 | <0.001 |
| 20–400 (n=66) | 27 | 39 | ||
| ≥400 (n=65) | 13 | 52 | ||
| Liver fibrosis | Established cirrhosis (n=71) | 39 | 32 | 0.875 |
| Portal fibrosis (n=31) | 15 | 16 | ||
| Fibrous speta (n=28) | 11 | 17 | ||
| Nodular formation and incomplete cirrhosis (n=9) | 3 | 6 | ||
| No Fibrosis (n=75) | 39 | 36 | ||
| Child Pugh classification grade | A (n=218) | 108 | 110 | 0.642 |
| B (n=21) | 11 | 10 | ||
| C (n=1) | 1 | 0 | ||
| relative number | ≤1 (n=65) | 31 | 34 | 0.321 |
| 2 (n=29) | 12 | 17 | ||
| 3 (n=15) | 11 | 4 | ||
| 4 (n=4) | 3 | 1 | ||
| ≥5 (n=3) | 1 | 2 | ||
| Person neoplasm status | With tumor (n=232) | 113 | 119 | 0.493 |
| Tumor free (n=114) | 60 | 54 | ||
| Vascular invasion | Macro (n=16) | 5 | 11 | 0.458 |
| Micro (n=94) | 48 | 46 | ||
| None (n=207) | 105 | 102 |
AJCC, American Joint Committee on Cancer; AFP, alpha fetal protein.
Significant genes and pathways identified by GSEA
AQP9 mRNA expression data for 374 samples in TCGA dataset were downloaded to clarify the biological role of co-regulated proteins by GSEA. We found that AQP9 inhibits some pathways, including the KRAS signaling pathway, the p53 pathway, hypoxia response, and TNF-α signaling pathway and some cancer signaling pathways (Figure 6A). The bubble chart revealed that AQP9 plays an important role as a tumor suppressor gene in HCC (Figure 6B).

Functional annotation and signaling pathways predicted by LinkedOmics
In this study, we also used LinkedOmics to detect AQP9 co-expressed genes in the LIHC cohort. All significant genes are shown in Figure 7A. The top 50 significant genes that correlated with AQP9 expression are shown in a heat map (Figure 7B). Significant GO term annotation by GSEA showed that the co-expressed genes participate in fatty acid metabolic process, lipid catabolic process, acute inflammatory response, and protein-lipid complex, while activities such as cell cycle G2/M phase transition, condensed chromosome, and Notch-binding were inhibited. KEGG) pathway analysis showed enrichment in retinol metabolism and drug metabolism (Figure 7C).
Prediction of target miRNA pathways
MicroRNAs (MiRNAs), which are widely distributed in eukaryotes, contain 21 to 23 nucleotides, and can regulate gene expression (26) by inhibiting binding specifically to target messenger RNAs (mRNAs) to inhibit their expression. MiRNAs play import roles in processes associated with tumor progression, such as proliferation, invasion, and angiogenesis (27). In this study, we investigated the ability of miRNAs to inhibit AQP9 expression in HCC. The TargetScan, miRDB, StarBase and mirDIP databases were used to predict interactions between AQP9 and miRNAs. Three candidate miRNAs were identified (Figure 8A), among which miR-23a-3p and miR-330-3p have been reported to promote the progression of liver cancer (28-30). We found that miR-23a-3p and miR-330-3p were overexpressed in HCC cell lines (Figure 8B,C), and the levels were inversely proportional to the expression of AQP9 in HCC tissues (Figure 8D,E).These findings suggested that the decreased expression of AQP9 in HCC may be due to the increased levels of miR-23a-3p and miR-330-3p.
AQP9 is associated with immune cell infiltration in HCC
Analysis of the TIMER database showed that AQP9 expression is significantly correlated with tumor purity (r=0.17, P=1.49×10−3) and the levels of infiltration by the immune cells types. In addition, the AQP9 expression levels in HCC tissues correlated with tumor infiltration by B cells (r=−0.289, P=8.83×10−8), CD8+ T cells (r=−0.295, P=2.76×10−8), CD4+ T cells (r=−0.254, P=1.85×10−6), macrophages (r=−0.369, P=2.07×10−12), neutrophils (r=−0.254, P=1.78×10−6) and DCs (r=−0.311, P=4.80×10−9) (Figure 9A). We also found that the expression levels of most marker sets of monocytes (Figure 9B), tumor-associated macrophages (Figure 9C), and M1 macrophages (Figure 9D) showed strong correlations with AQP9 expression, although M2 macrophages did not (Figure 9E). Therefore, our data confirmed that AQP9 is correlated with immune infiltrating cells and suggested that AQP9 plays an important role in the liver cancer microenvironment.
Discussion
Liver cancer has a high mortality rate worldwide. Despite the availability of some effective treatments in the clinic, the OS of patients is still unsatisfactory (1). Most patients show no typical symptoms in the early stages of liver cancer. AFP is frequently used as an indicator for early screening of HCC frequently, although less than 70% of liver cancer patients are AFP-positive (31).
With changes in lifestyle and eating habits, the incidence of NAFLD has increased. In addition, effective vaccination strategies against hepatitis viruses have become more acceptable, and the etiology of liver diseases has altered (10). Considering these factors, new markers of HCC are needed to improve early diagnosis.
The mechanism of tumorigenesis is still unclear. The genetic factors, immune status and external mutagens are the basis of, and conditions for tumorigenesis. Chemical and physical factors and virus infection can activate proto-oncogenes, inactivate tumor suppressor genes, and rearrange or mutate genes, which eventually lead to abnormal gene expression and tumorigenesis (32). AQPs have been reported to be involved in tumor progression (7,8,33). Among them, AQP9 regulates the water and glycerol permeability. In humans, AQP9 is an essential aquaporin in the liver, researchers find that overexpress AQP9 may reduce the levels of PCNA, E-cad, N-cad, α-SMA, DVL2, GSK-3β, cyclinD1 and β-catenin in HCC cells, and cell apoptosis was remarkably enhanced. In addition, its over-expression suppresses cell invasion through inhibiting epithelial-to-mesenchymal transition. AQP9 acts as a tumor suppressor in HCC invasion via the regulation of HIF-1α expression in the tumor hypoxic microenvironment. Furthermore, AQP9 overexpression was found to result in reactive oxygen species (ROS) accumulation, which inhibited β-catenin activity by attenuating the interaction of β-catenin with TCF4 while concurrently enhancing the association of β-catenin with FOXO3a, ultimately inhibiting liver cancer stem cells (LCSCs) (12,14-17); However, the prognostic value of AQP9 and the specific mechanism underlying its role in HCC remain to be further elucidated. Therefore, in this study, we conducted a bioinformatics analysis of AQP9 in liver cancer.
Database analysis revealed that AQP9 is expression is significantly decreased in liver cancer compared with the high expression levels in normal liver. Furthermore, RT-qPCR, Western blot and immunohistochemistry analysis of our samples also indicated that AQP9 is downregulated in HCC. Moreover, AQP9 was negatively correlated with different stage, grade, weight, lymph node metastasis, sex, and histological subtypes. We also found that AQP9 has great diagnostic value for HCC, with low levels of AQP9 linked to worse OS, and different stages and grades. Analysis of clinical data downloaded from TCGA showed significant differences in AQP9 expression in AJCC tumor stage (P=0.018), age (P=0.0013), sex (P<0.001) and AFP (P<0.001); therefore, we propose that AQP9 is a valuable diagnostic and prognostic marker in liver cancer.
GSEA and functional enrichment analysis showed that AQP9 was significantly involved in the major hallmark pathways. AQP9 inhibits some cancer-related pathways, such as hypoxia response, and some studies have shown that AQP9 overexpression leads to reactive oxygen species (ROS) accumulation, and inhibition of β-catenin activity (16), which in turn, leads to decreased HIF-1α expression (17). AQP9 was also significantly involved in p53, G2M checkpoint, and the KRAS, and TNF-α-NFκB signaling pathways, although further studies are required to clarify these associations. Among the enrichment pathways related to AQP9, most were inhibited, and these pathways are related to carcinogenesis. We also used LinkedOmics to investigate the network of AQP9 and its most closely related genes. We found some genes that were related to fat production and metabolism, and others were significantly involved in gene ontology and KEGG pathways. All these findings suggest a role for AQP9 as a tumor suppressor gene in liver cancer.
AQP9 expression is downregulated in HCC, but the cause of this decrease is unclear. Under some conditions, miRNAs regulate target genes and downstream signaling pathways. Many studies have shown that miRNAs are important in tumor progression (34). It has been confirmed that miRNAs regulate target genes to participate in the progression of liver cancer. In particular, miR-21 contributes to HCC growth and modulation of PTEN expression (35), and miR-330-5p targets SPRY2 and activates MAPK/ERK signaling to promoting HCC progression (36). In this study, we investigated the potential role of miRNAs in the decreased expression of AQP9 in HCC. We used the TargetScan, miRDB, StarBase and mirDIP databases to predict miRNAs that target AQP9. Among the three candidates, miR-23a-3p and miR-330-3p have been reported in HCC (28-30). Our data show that mir-23a-3p and mir-330-3p are expressed in HCC cell lines, and our analysis of the datasets in the databases indicates that AQP9 expression is inversely proportional to mir-23a-3p and mir-330-3p in HCC tissues. Therefore, we hypothesize that mir-23a-3p and mir-330-3p inhibit the expression of AQP9 in HCC, although further studies are required to confirm this.
Tumor immunotherapy is a new strategy, which has received increasing attention in relation to HCC (37). This is a potential field of research into the tumor microenvironment for biomarkers or therapeutic targets for prognosis and diagnosis of HCC (38). Immune cell infiltration has a great influence on survival in HCC patients (39,40). We identified a significantly positive correlation between AQP9 expression and the level of infiltration by B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and DCs. The expression levels of some marker set of monocytes, TAMs, and M1 macrophages also showed strong correlations with AQP9 in HCC.
Conclusions
In this study, we conducted a detailed analysis of AQP9 expression and explored its relationship with the prognosis of HCC patients. Our results indicated that AQP9 was significantly decreased in HCC, with low AQP9 levels indicating a poor outcome. GSEA analysis and LinkedOmics revealed that AQP9 was significantly involved in the major hallmark pathways. Mechanistically, mir-23a-3p and mir-330-3p may inhibit AQP9 expression in HCC. Finally, our results suggest that AQP9 may be a target for immunotherapy of liver cancer. The incidence of NAFLD is now increasing, and our results indicate that AQP9, which is a water-glycerol channel protein, may represent a potential therapeutic target in HCC.
Acknowledgments
Thanks LEXIS (Scientific Editing Experts, United States. LEXIS Academic Service, LLC) helps us polish our manuscript.
Funding: This study was supported by grants from
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-3158
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-3158
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3158). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from the patients, and this research was approved by the Ethics Committee of Affiliated Hospital of Nantong University (ethical code: 2018-L006).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021;73:4-13. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-24. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-91.e1. [Crossref] [PubMed]
- Agre P, Preston GM, Smith BL, et al. Aquaporin CHIP: the archetypal molecular water channel. Am J Physiol 1993;265:F463-76. [PubMed]
- Li C, Wang W. Molecular Biology of Aquaporins. Adv Exp Med Biol 2017;969:1-34. [Crossref] [PubMed]
- Wen J, Wang Y, Gao C, et al. Helicobacter pylori infection promotes Aquaporin 3 expression via the ROS-HIF-1alpha-AQP3-ROS loop in stomach mucosa: a potential novel mechanism for cancer pathogenesis. Oncogene 2018;37:3549-61. [Crossref] [PubMed]
- Dai C, Charlestin V, Wang M, et al. Aquaporin-7 Regulates the Response to Cellular Stress in Breast Cancer. Cancer Res 2020;80:4071-86. [Crossref] [PubMed]
- Chetry M, Li S, Liu H, et al. Prognostic values of aquaporins mRNA expression in human ovarian cancer. Biosci Rep 2018;38:BSR20180108. [Crossref] [PubMed]
- Zhou J, Zhou F, Wang W, et al. Epidemiological Features of NAFLD From 1999 to 2018 in China. Hepatology 2020;71:1851-64. [Crossref] [PubMed]
- Gregoire F, Lucidi V, Zerrad-Saadi A, et al. Analysis of aquaporin expression in liver with a focus on hepatocytes. Histochem Cell Biol 2015;144:347-63. [Crossref] [PubMed]
- Gena P, Mastrodonato M, Portincasa P, et al. Liver glycerol permeability and aquaporin-9 are dysregulated in a murine model of Non-Alcoholic Fatty Liver Disease. PLoS One 2013;8:e78139. [Crossref] [PubMed]
- Cai C, Wang C, Ji W, et al. Knockdown of hepatic aquaglyceroporin-9 alleviates high fat diet-induced non-alcoholic fatty liver disease in rats. Int Immunopharmacol 2013;15:550-56. [Crossref] [PubMed]
- Liao S, Chen H, Liu M, et al. Aquaporin 9 inhibits growth and metastasis of hepatocellular carcinoma cells via Wnt/beta-catenin pathway. Aging (Albany NY) 2020;12:1527-44. [Crossref] [PubMed]
- Zhang WG, Li CF, Liu M, et al. Aquaporin 9 is down-regulated in hepatocellular carcinoma and its over-expression suppresses hepatoma cell invasion through inhibiting epithelial-to-mesenchymal transition. Cancer Lett 2016;378:111-19. [Crossref] [PubMed]
- Zheng X, Li C, Yu K, et al. Aquaporin-9, Mediated by IGF2, Suppresses Liver Cancer Stem Cell Properties via Augmenting ROS/beta-Catenin/FOXO3a Signaling. Mol Cancer Res 2020;18:992-03. [Crossref] [PubMed]
- Qian Y, Liu F, Zhang W, et al. AQP9 suppresses hepatocellular carcinoma cell invasion through inhibition of hypoxia-inducible factor 1alpha expression under hypoxia. J Gastroenterol Hepatol 2020;35:1990-97. [Crossref] [PubMed]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci 2018;27:233-44. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya S, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-04. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-07. [Crossref] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [Crossref] [PubMed]
- Liu J, Fan L, Yu H, et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019;70:241-58. [Crossref] [PubMed]
- Zhao X, Chen GQ, Cao GM. Abnormal expression and mechanism of miR-330-3p/BTG1 axis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2019;23:6888-98. [PubMed]
- Xiang Y, Yang Y, Lin C, et al. MiR-23a-3p promoted G1/S cell cycle transition by targeting protocadherin17 in hepatocellular carcinoma. J Physiol Biochem 2020;76:123-34. [Crossref] [PubMed]
- Luo P, Yin P, Hua R, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 2018;67:662-75. [Crossref] [PubMed]
- Wang LH, Wu CF, Rajasekaran N, et al. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell Physiol Biochem 2018;51:2647-93. [Crossref] [PubMed]
- Wu Q, Yang ZF, Wang KJ, et al. AQP8 inhibits colorectal cancer growth and metastasis by down-regulating PI3K/AKT signaling and PCDH7 expression. Am J Cancer Res 2018;8:266-79. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647-58. [Crossref] [PubMed]
- Xiao S, Yang M, Yang H, et al. miR-330-5p targets SPRY2 to promote hepatocellular carcinoma progression via MAPK/ERK signaling. Oncogenesis 2018;7:90. [Crossref] [PubMed]
- El DI, Khalil DN, Abou-Alfa GK. Immune checkpoint inhibitors for hepatocellular carcinoma. Cancer 2019;125:3312-19. [Crossref] [PubMed]
- Hilmi M, Vienot A, Rousseau B, et al. Immune Therapy for Liver Cancers. Cancers (Basel) 2019;12:77. [Crossref] [PubMed]
- Huang CY, Wang Y, Luo GY, et al. Relationship Between PD-L1 Expression and CD8+ T-cell Immune Responses in Hepatocellular Carcinoma. J Immunother 2017;40:323-33. [Crossref] [PubMed]
- Zheng C, Zheng L, Yoo JK, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342-56.e16. [Crossref] [PubMed]


