
A microarray study of radiation-induced transcriptional responses and the role of Jagged 1 in two closely-related lung cancer cell lines
Introduction
Lung cancer is the leading cause of cancer-related deaths globally (1). According to a report of the American Cancer Society (ACS) that was published in 2015, lung cancer is the most common cancer in both males and females, accounting for 28% and 26% of their cancer-caused deaths, respectively (2). Additionally, of all of the deaths that are caused by malignant tumors, lung cancer caused more than any other in Taiwan. Lung cancer is classified as small cell lung cancer, adenocarcinoma (the predominant type), squamous cell carcinoma or large cell carcinoma, based on cell morphology (3). At presentation, the cancers in more than one-third of patients with adenocarcinoma, squamous cell carcinoma or large cell carcinoma are in locally advanced stage IIIA or IIIB, and patients with cancers in these stages are suitable for radiotherapy rather than surgical resection. Unfortunately, radiotherapy alone or in combination with chemotherapy does not significantly improve 5-year survival rates, which reach only 10-15% (4,5).
Ionizing radiation causes many types of DNA damage, including DNA double strand breaks, and various cellular responses that protect the cells from damage (6). The cellular responses include cell cycle arrest, apoptosis, DNA repair and activated signaling pathways (7,8). The cell cycle checkpoints respond to ionizing radiation, delaying the normal progression of the cell cycle, providing time for DNA repair-related genes to facilitate DNA repair. The phase of G2/M arrest with cells is considered to be both a passive response of DNA damage and an active response to promote DNA repair (9-11), and this process is mediated by various mediators and effectors. However, if the cell cycle arrest response does not occur, then the cells undergo apoptosis or mitotic death owing to the failure to repair DNA (12).
Jagged 1, a Notch receptor ligand, has an important role in the progression of various human cancers. In human prostate cancer, the up-regulation of Jagged 1 protein occurs in metastatic tumors relative to that its expression in localized tumors or benign tissues, indicating that Jagged 1 may be a potential biomarker that can distinguish indolent from aggressive prostate cancers (13). In breast cancer, high expressions of Jagged 1 mRNA and protein predict a poor prognostic outcome, and can be used to identify patients that will benefit from therapy or not (14). In squamous cell lung carcinoma and carcinoid tumors, Jagged 1 shows significantly different expressions (15). A previous study revealed that the unaffected parents of retinoblastoma (Rb) patients were hypersensitive to radiation, and Jagged 1 exhibited a significant change in expression indicating that it may play a role in radio-sensitivity (16). Additionally, the activated Notch pathway contributes to the radio-resistance of gliomas stem cells (17). Therefore, Jagged 1 may be associated with radio-sensitivity.
The mechanism by which lung cancer responds to radiation is still unclear. Therefore, the need to understand the effects of radiation on lung cancer is urgent. The molecular biomarkers of radiation responses could elucidate the process of radiotherapy, and potentially predict the treatment outcome of individuals. In this study, lung adenocarcinoma cell lines, CL1-0 and CL1-5, which have different metastasis capacities were found, by clonogenic assay, to exhibit different degrees of radiation-induced cyto-toxicity and were used to identify differentially expressed genes that are involved in the different radiation responses using microarray technology. From the array data, the expression of Jagged 1 differed considerably between these two cell lines. Finally, irradiation reduced the surviving fraction of Jagged 1-overexpressed CL1-0 cells, indicating that Jagged 1 has a role in the radio-sensitivity of CL1-0 and CL1-5 cell lines.
Materials and methods
Cell culture
Human lung cancer cell lines with differential invasiveness, CL1-0 and CL1-5, were obtained from Dr. Pan-Chyr Yang’s group. CL1-0 and CL1-5 cells were maintained in RPMI-1640 (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (Biological industries, Israel) and were grown exponentially at 37 °C in 5% CO2 (18,19). The cell lines were subcultured approximately two to three times weekly. The cells were counted using a Z2 Coulter Particle Counter (Beckman Coulter, CA, USA).
Radiation treatment
Cells with 70-80% confluences in a medium of height 0.5 cm were grown in 10 cm petri dishes and irradiated with a cobalt 60 (Picker V9) source at dose rates between 270 and 330 cGy/min.
Clonogenic assay
Various amounts of untreated cells or irradiated cells were seeded into 10 cm dishes. After two weeks of incubation, the cells were prepared for counting under microscope; they were firstly washed in PBS, fixed with methanol and acetic acid (3:1, v/v) for 1 hour (hr), and then stained with crystal violet (0.1%) for 1 hr.
Flow cytometric analysis and quantification of apoptosis
The attached cells were trypsinized and re-suspended in medium. After centrifugation at 200 g for 10 min, the medium was removed and the cell pellets were washed twice in PBS. They were then resuspended in 500 µL PBS and 500 µL methanol, before being centrifuged at 200 g for 5 min. After centrifugation, the cell pellets were fixed in 1 mL methanol and stored at −20 °C. For analysis, the cells were collected by centrifugation at 200 g for 5 min and resuspended in PBS that contained 10 μg/mL propadium iodide (PI), 0.2 mg/mL RNase A, and 0.1% Triton X-100. The mixture was allowed to react in the dark for 30 minutes. The cell suspension mixtures were then filtered and analyzed by FACSCalibur flow cytometry (BD Bioscience, USA) and the CellQuest program was used to determine DNA content and cell cycle distribution.
RNA preparation
At each collection point (no radiation treatment, 0 hr; after exposure to 10 Gy radiation for 1, 4 and 24 hr), the cells were washed twice in ice cold PBS, and then scraped off using 1 mL Tri Reagent® (Molecular Research Center, Cincinnati, USA). Cells in Tri Reagent® were incubated at 4 °C for 10 minutes. Then, 100 µL of BCP (1-bromo-3-chloropropane (Molecular Research Center, Inc.) was added to every 1 mL of Tri Reagent® and then vortexing was performed. After 10 minutes of reaction on ice, the mixtures were centrifuged at 13,200 rpm at 4 °C for 10 min; the supernatant was collected and mixed with isopropanol and then stored at −80 °C for 2 hrs to allow RNA precipitation. After 2 hours, the samples were centrifuged at 13,200 rpm at 4 °C for 30 min, before being washed twice with 70% ethanol. Finally, the RNA pellets were resuspended in 100 µL nuclease-free water. Total RNA was purified using the RNeasy® Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions.
Whole genome gene expression
For Illumina BeadChip microarray experiment, 500 ng of total RNA was amplified using Illumina Total Prep RNA amplification kit (Ambion, Inc., USA). RNA samples were reverse-transcribed using a T7 oligo (dT) primer and then synthesized second-strand cDNA. The double-stranded cDNA templates were then used to generate biotinylated cRNA by in vitro transcription (IVT) for 14 hr. Then, 0.75 µg cRNA was applied to an Illumina Human Ref-8 V3 BeadChip (Illumina, Inc., USA) to hybridize for 16 hrs. After overnight hybridization, the beadchip was washed, stained with streptacidin-Cy3 (GE Healthcare Bio-Sciences), and scanned by Illumina BeadStation. The images thus obtained were analyzed by Bead Studio (Illumina, Inc.).
Data analysis
The intensity result files from the microarray experiment were analyzed by Partek (St. Louis, MO, USA) to quantify mRNA expression. Differentially expressed genes that exhibited a difference in expression of at least 1.8-fold between the untreated and radiation treated samples were selected. Tight hierarchical clustering (20) was used further to group genes with similar expression profiles. Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA, USA) was used in the functional analysis of the selected genes.
Quantitative real-time PCR
The genes that were identified from the microarray analyses were verified by quantitative real-time PCR assay. Quantitative real-time PCR was performed by taking 1 µg of total RNA that was extracted from CL1-0 and CL1-5. Total RNA was reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The reaction mixture was incubated at 25 °C for 10 min, at 37 °C for 2 hr and at 85 °C for 5 sec. Real-time PCR was performed using the SYBR Green PCR Master Mix in the 7300 real-time PCR System (Applied Biosystems). The reactions were carried out with the following program; 40 cycles of denaturing at 95 °C for 15 sec and annealing and elongating at 60 °C for 1 min. Each measurement was made in triplicate and normalized to GAPDH as the internal control. The sequences of real-time PCR primers were as follows.
Jagged 1: 5'-TTGTGAGCCTAATCCCTGCCAGAA-3' (forward), 5'-AGTGGTCTTTCAGGTGTGAGCAGT-3' (reverse);
NFκBIA: 5'-AGACCTGGCTTTCCTCAACTTCCA-3' (forward), 5'-TCAGCAATTTCTGGCTGGTTGGTG-3' (reverse);
NFκBIZ: 5'-TAGATGCTGTCCGCCTGTTGATGA-3' (forward), 5'-CCAAATGCACTGGCTGTTCGTTCT-3' (reverse);
GAPDH: 5'-CCCTCAACGACCACTTTGTC-3' (forward), 5'-CTTACTCCTTGGAGGCCATG-3' (reverse).
Construction of overexpression vector
Fagged 1 was cloned into the expression vector pcDNA3.1. The pcDNA3.1-JAG1 vector was then fully sequenced to ensure that no mutations had been introduced during the PCR amplification.
Western blotting analysis
Cells were washed twice with ice-cold PBS and then collected at the different time points (1, 4, 24 hr) after exposure to 10 Gy radiation. The cell pellets were then lysed using a lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 5 mM EDTA, 0.02 mM EGTA, and 1% protease inhibitor cocktail) by incubating at 4 °C for 25 min. After centrifugation, the protein concentrations of the supernatants were determined using a BCA assay kit (Pierce), and 50 µg of each sample was loaded onto the 8% SDS polyacrylamide gel. After electrophoresis, the proteins were transferred onto a PVDF membrane. The membrane was blocked in TBST/5% skimmed milk and then incubated with primary antibody (Anti-Jagged1, BD Bioscience, USA, 130 kDa) in TBST/5% skimmed milk. The membrane was then washed three times with TBST, before incubation with secondary antibody in TBST/5% skimmed milk. The signals were detected by Immobilon Western (Millipore, Billerica, MA, USA), and the images were captured by the BioSpectrum Imaging System (UVP, Upland, CA, USA).
Results
Clonogenic assay and cell cycle profile analysis of CL1-0 and CL1-5
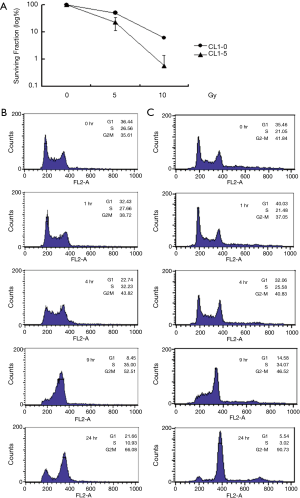
To determine the ability of the cells to form colonies after radiation treatment, CL1-0 and CL1-5 cells were irradiated with γ-rays at doses of 5, 10, and 20 Gy. Exposure of CL1-0 and CL1-5 cells to 5 Gy radiation reduced their survival rates to 49.48% and 22.95%, respectively; exposure to 10 Gy radiation reduced them to 6.08% and 0.57%, respectively (Figure 1A). However, exposure to 20 Gy radiation prevented the formation of any colony of either cell line (data not shown). The survival rate of CL1-0 was higher than that of CL1-5 at both doses of radiation, and upon exposure to 10 Gy radiation, the difference between the survival rates of the two cell lines differed by an order of magnitude. The results revealed that CL1-0 was more radio-resistant than CL1-5. The cell cycle distribution results showed that the G2/M percentage increased slowly with time for both CL1-0 and CL1-5 cell lines (Figure 1B,C). The declining cell populations in the G1 and S phases and the increasing population in the G2/M phase showed the same cell cycle progression in these two cell lines. However, after exposure to radiation, the proportion of the CL1-5 cell population in the G2/M phase considerably exceeded of the CL1-0 population.
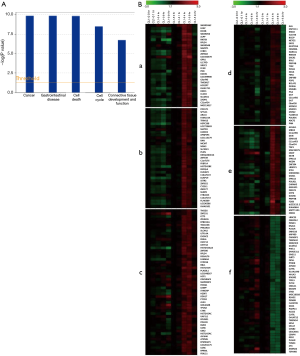
Identification of differentially expressed genes and gene expression profiles
In the Illumina microarray experiment, genes in CL1-0 and CL1-5 which expressions were at least 1.8 times than those in the untreated 0 hr samples were selected. From the array data, 1,595 genes were identified as differentially expressed in the two cell lines at one or more time points. The main molecular and cellular functions of these 1,595 genes were further analyzed by IPA and related to cell death, cell cycle, cell growth and proliferation, cellular function and maintenance (Figure 2A). To examine the global expression profiles of these 1,595 differentially expressed genes, they underwent tight hierarchical clustering to observe the similarities in their expression patterns over time. In six clusters, the expression profiles of CL1-0 and CL1-5 differed significantly, possibly explaining the different radio-sensitivities at the transcriptional level (Figure 2B). Tables S1-S6 present the genes in the six clusters. Genes in clusters a, b and c presented in similarly, indicating that these genes in CL1-5 were activated at 4 to 9 hr post-irradiation, and their expression levels declined at 24 hr. However, in CL1-0, the expression of these genes was either repressed or remained unchanged throughout the time course. Genes in clusters d and e in the CL1-5 were up-regulated at 4 and 9 hr, and down-regulated at 24 hr; in CL1-0, their expression remained unchanged or repressed throughout the time course. Genes in cluster f in CL1-5 showed down-regulation at 4 and 9 hr, but those in CL1-0 were unchanged at 4hr then were up-regulated at 24 hr.
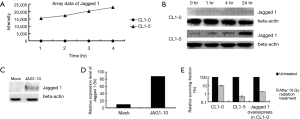
Analyses of Jagged 1 expression after exposure to radiation and its effect on cellular survival
According to the microarray data, the Jagged 1 gene expression level in CL1-5 markedly exceeded that in CL1-0 either before or after 10 Gy radiation treatment (Figure 3A). Meanwhile, the Jagged 1 protein expression level increased with duration of 10 Gy radiation treatment in CL1-5 and the increases at 1, 4, 24 hr were 2.65-fold, 4.15-fold, and 5.30-fold respectively, relative to the untreated sample (0 hr). In CL1-0, Jagged 1 protein expressions were lower than in CL1-5 at all time points (Figure 3B). These results demonstrated that the transcriptional and translational expression patterns of Jagged 1 in CL1-0 and CL1-5 upon treatment with 10 Gy radiation were matched to each other. To verify the role of Jagged 1 in determining the responses of lung cancer cells to radiation, Jagged 1 was overexpressed in the CL1-0 cells. The expression level of Jagged 1 in CL1-0 was verified by real-time PCR and western blotting. The results clearly demonstrated that the Jagged 1 expression level in the overexpressed CL1-0 cells was about three-fold higher than that in the mock cells (Figure 3C,D). The cell survival rates of CL1-0, CL1-5, and Jagged 1-overexpressed CL1-0 cells following treatment with 10 Gy radiation were measured (Figure 3E) and found to be reduced to 9.24%, 0.46%, and 1.79%, respectively. The survival rate of Jagged 1-overexpressed CL1-0 was significantly lower than that of CL1-0 (P<0.05), indicating that Jagged 1 may play a role in influencing the radio-sensitivity of lung cancer cells.
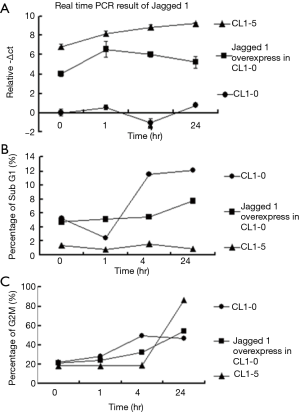
Alterations of cell cycle profile caused by Jagged 1 over-expression in CL1-0 cells
Since Jagged 1 may be involved in radio-sensitivity, the cell cycle profiles of the Jagged 1 overexpressed CL1-0 cells were further examined. The real-time results showed that the expression of Jagged 1 increased slowly over time in CL1-5 and overexpressed CL1-0, and fluctuated around a much lower value in CL1-0 (Figure 4A). To investigate the effect of radiation treatment on Jagged 1-overexpressed CL1-0, the subpopulation before G1 (Sub G1) and the cell cycle distribution were determined. The results thus obtained reveal that the amount of sub G1 in Jagged 1-overexpressed CL1-0 at 24 hr was between that in CL1-0 and that in CL1-5 (Figure 4B), and similar results were observed in relation to the percentage of G2/M (Figure 4C). Taken together, the percentages of sub G1 and G2/M overexpressed CL1-0 at 24 hr were lower than those of CL1-0, indicating that overexpressed Jagged 1 in CL1-0 may affect the cell cycle distribution.
Discussion
Mechanisms of cell survival of CL1-0 and CL1-5 cells
A significant difference between the cell survival of CL1-0 and that of CL1-5 was observed following radiation treatment. The cell survival of CL1-5 was approximately one tenth that of CL1-0. Accordingly, the differentially expressed genes were not only the genes that responded to radiation, but also those related to cell viability. As expected, in both cell lines, several genes related to cell signaling, survival, and DNA repair exhibited different expression levels following treatment with 10 Gy radiation. The genes in the six clusters were differently expressed between CL1-0 and CL1-5, possibly explaining the different cell survival rates. Twenty-eight genes were sub-grouped into cluster a. XRCC2 in cluster a participates in homologous recombination (HR) to maintain chromosome stability and repairs DNA damage (21). Prohibitin (PHB) negatively regulates cell proliferation, and is suggested to be a tumor suppressor that is associated with the immune response. In cluster b with 35 genes, the cancer-associated network includes members of the RAS oncogene family (RAN), which is involved in cell cycle progression and DNA synthesis. In the carbohydrate metabolism associated network classification, protein O-fucosyltransferase 1 (POFUT1) participates in ligand-induced receptor signaling, including Notch signaling in mammals (22). In cluster c with 51 genes, gene expression associated network classification includes BUB3, which is involved in the spindle checkpoint function. Centromere protein F (CENPF) is associated with the centromere-kinetochore complex and plays a role in chromosome segregation during mitosis (G2/M phase). The distribution of cells in the G2/M phase in CL1-0 and CL1-5 can be verified with reference to BUB3 and CENPF gene expression patterns. In cluster d with 35 genes, the cell cycle associated network classification includes mini chromosome maintenance complex component 10 (MCM10) and mini chromosome maintenance complex component 2 (MCM2), which are related to the initiation of eukaryotic genome replication. BRCA2 and CDKN1A interacting protein (BCCIP) is a (cofactor for BRCA2 and a modulator of CDK2 kinase activity. BCL2-associated transcription factor 1 (BCLAF1), which is related to apoptosis, can be suppressed by the co-expression of BCL2 proteins. In cluster e, cAMP responsive element binding protein 1 (CREB1) participates in encoding a transcription factor that is a member of the leucine zipper family of DNA binding proteins. In the protein synthesis- and cell death-associated network classification, gene interferon gamma receptor 2 (IFNGR2) helps to encode the non-ligand-binding beta chain of the gamma interferon receptor. Cyclin B2 (CCNB2) is involved in G2/M DNA damage checkpoint regulation.
Data on cell cycle distribution may indicate that CL1-0 and CL1-5 cells die by different mechanisms after 10 Gy radiation treatment. Since CL1-0 and CL1-5 are p53-mutated cell lines, the G1/S checkpoint was absent and cells were arrested in the G2/M phase with cumulatively damaged DNA. Double strand breaks can be repaired by HR during G2/M arrest (23). In this study, cells with excessive chromosome lesions might have three results: they directly bypassed the G2/M checkpoint to begin endoreduplication cycles from G2 arrest, or they were subsequently detected by the spindle checkpoint and presented with abortive mitosis or mitotic death. More CL1-5 cells than CL1-0 cells were in the G2/M phase at 24 hr. Moreover, a small peak were detected at around 8N reveals that little amount of CL1-5 cells passed a G2/M checkpoint and then into an endoreduplication cycle. Therefore, the major survival mode of CL1-5 may involve initiating endocycles during G2/M arrest, delaying death. In CL1-0, of which a smaller proportion was in the G2/M phase at 24 hr, revealed that CL1-0 passed a G2/M checkpoint and then, in most cases, apoptosis, rather than entering the endoreduplication cycle. However, the data of cell cycle distribution was only collected within 24 hrs following irradiation and a longer period time of observation was needed to explain the result of clonogenic assay.
Jagged 1-related cell survival mechanisms in CL1-0 and CL1-5
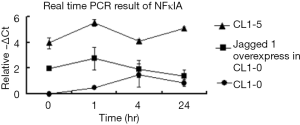
In this study, Jagged 1 exhibited consistent transcriptional and translational expression profiles in CL1-0 and CL1-5 that were untreated or treated with 10 Gy radiation. Following radiation treatment, the cell population of Jagged 1-overexpressed CL1-0 in the sub G1 and G2/M phases and the survival rate thereof, were between those of CL1-0 and CL1-5. These results suggested that Jagged 1 influences radio-sensitivity. Since Jagged 1 is an important ligand in the Notch pathway that is related to cell proliferation and apoptosis, Jagged 1-overexpressed CL1-0 was expected to exhibit a different degree of radio-sensitivity from that of the original CL1-0 cells. Moreover, collaboration of Notch and NFkB signaling is reportedly important in the proliferation, differentiation and apoptosis of a diverse range of cells (24). Expression levels of NFκBIA in CL1-0, Jagged 1-overexpressed CL1-0, and CL1-5 were also measured with real-time PCR (Figure S1). Up-regulation was followed by the suppression of NFκBIA in Jagged 1-overexpressed CL1-0, and this pattern of expression differed from that of CL1-5. In CL1-5, the expression level of NFκBIA was recovered at 24 hr, rather than remaining continuously repressed, as in Jagged 1-overexpressed CL1-0. This finding may explain the higher survival rate of Jagged 1-overexpressed CL1-0 than that of CL1-5. The different expression patterns of NFκBIA in the Jagged 1-overexpressed CL1-0 and CL1-0 indicated that the expression level of Jagged 1 may influence the NFκBIA patterns.
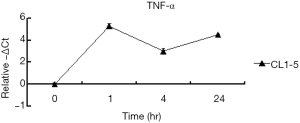
To identify the signal in response to the radiation treatment of CL1-0 and CL1-5, the level of tumor necrosis factor alpha (TNF-α) was measured by real-time PCR because TNF-α is a very common radio-inducible cytokine (25). The results thus obtained showed an up-regulation at 1 hr following 10 Gy radiation treatment in CL1-5, similar to the pattern of expression of NFκBIA in CL1-5 (Figure S2). A previous study found that high-dose irradiation induces a significant release of TNF-α in the lung adenocarcinoma. TNF-α can activate the transport of NFκB into the nucleus, resulting in the activation of NFκB signaling. Furthermore, the survival of lung adenocarcinoma has been reversed using a neutralizing antibody against TNF-α (25). Therefore, TNF-α may provide the death signal in CL1-5 following treatment with 10 Gy radiation.
In summary, radio-sensitivity of two lung adenocarcinoma cell lines, CL1-0 and CL1-5, was investigated in this study. Clonogenic assay results showed that CL1-5 was more radio-sensitive than CL1-0. The proportion of cell population at G2/M phase in CL1-5 was markedly higher than that in CL1-0 after 24 hours of radiation exposure. Moreover, a total of 1,595 differentially expressed genes were identified between these two lung cancer cell lines using gene expression microarrays. Hierarchical clustering analysis of the differentially expressed genes highlighted six clusters of genes and their functional associations with cell cycle, cell death and DNA replication. Jagged 1, a Notch ligand, was found to be significantly overexpressed in CL1-5 after 10 Gy radiation treatment. Functional assay results showing a decreased cell survival rate of Jagged 1-overexpressed CL1-0 after radiation exposure as compared with that of CL1-0 further suggest that Jagged 1 may play a role enhancing the radio-sensitivity of lung cancer. The finding described in this work may potentially facilitate the development of new radiation treatments for lung adenocarcinoma.
Supplementary materials
Six clusters of differentially expressed genes and their related biological networks (Tables S1-S6)

Full table

Full table
Full table

Full table

Full table
Full table
Acknowledgments
Funding: The authors would like to thank the National Science Council of Taiwan for financially supporting this research under Contract No. MOST103-2314-B-002-034-MY3.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Lyudmila Bazhenova and Ajay Pal Singh Sandhu) for the series “Recent advances in radiotherapy and targeted therapies for lung cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2015.08.09). The series “Recent advances in radiotherapy and targeted therapies for lung cancer” was commissioned by the editorial office without any funding or sponsorship. EYC serves as the Editor-in-Chief of Translational Cancer Research. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study needs no approval of the Institutional Review Board (IRB) due to containing no samples from patients. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [PubMed]
- Travis WD. Pathology of lung cancer. Clin Chest Med 2002;23:65-81. viii. [PubMed]
- Perez CA, Pajak TF, Rubin P, et al. Long-term observations of the patterns of failure in patients with unresectable non-oat cell carcinoma of the lung treated with definitive radiotherapy. Report by the Radiation Therapy Oncology Group. Cancer 1987;59:1874-81. [PubMed]
- Chang GC, Chen KC, Yang TY, et al. Activity of gefitinib in advanced non-small-cell lung cancer with very poor performance status. Invest New Drugs 2005;23:73-7. [PubMed]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis 2002;23:687-96. [PubMed]
- Dent P, Yacoub A, Contessa J, et al. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res 2003;159:283-300. [PubMed]
- Iliakis G, Wang Y, Guan J, et al. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene 2003;22:5834-47. [PubMed]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science 1989;246:629-34. [PubMed]
- Lücke-Huhle C. Alpha-irradiation-induced G2 delay: a period of cell recovery. Radiat Res 1982;89:298-308. [PubMed]
- Tobey RA. Different drugs arrest cells at a number of distinct stages in G2. Nature 1975;254:245-7. [PubMed]
- Nias AH, editor. An introduction to radiobiology. 2nd. Chichester, England: John Wiley & Sons Ltd., 2000.
- Santagata S, Demichelis F, Riva A, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res 2004;64:6854-7. [PubMed]
- Dickson BC, Mulligan AM, Zhang H, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol 2007;20:685-93. [PubMed]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790-5. [PubMed]
- Fitzek MM, Dahlberg WK, Nagasawa H, et al. Unexpected sensitivity to radiation of fibroblasts from unaffected parents of children with hereditary retinoblastoma. Int J Cancer 2002;99:764-8. [PubMed]
- Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem cells 2010;28:17-28. [PubMed]
- Chu YW, Yang PC, Yang SC, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol 1997;17:353-60. [PubMed]
- Yang PC, Luh KT, Wu R, et al. Characterization of the mucin differentiation in human lung adenocarcinoma cell lines. Am J Respir Cell Mol Biol 1992;7:161-71. [PubMed]
- Tseng GC, Wong WH. Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics 2005;61:10-6. [PubMed]
- Mohindra A, Bolderson E, Stone J, et al. A tumour-derived mutant allele of XRCC2 preferentially suppresses homologous recombination at DNA replication forks. Hum Mol Genet 2004;13:203-12. [PubMed]
- Stahl M, Uemura K, Ge C, et al. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem 2008;283:13638-51. [PubMed]
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 2003;22:5792-812. [PubMed]
- Ang HL, Tergaonkar V. Notch and NFkappaB signaling pathways: Do they collaborate in normal vertebrate brain development and function? Bioessays 2007;29:1039-47. [PubMed]
- Shareef MM, Cui N, Burikhanov R, et al. Role of tumor necrosis factor-alpha and TRAIL in high-dose radiation-induced bystander signaling in lung adenocarcinoma. Cancer Res 2007;67:11811-20. [PubMed]


