
A comprehensive bioinformatics analysis to identify a candidate prognostic biomarker for ovarian cancer
Introduction
Ovarian cancer (OC) is a highly malignant tumor of the female reproductive system. Globally, there are nearly 240,000 new cases of OC diagnosed each year, and the death rate exceeds 50% (1,2). Patients with localized OC can be treated with standard treatments and have a good 5-year survival rate (3,4). However, detecting OC at an early stage is extremely challenging. Two-thirds of patients are diagnosed with an advanced stage of OC; in such cases, the disease has already spread throughout the peritoneal cavity and distantly metastasized at the time of diagnosis (5). Standard treatments have limited efficacy for advanced or recurrent OC, and the 5-year survival rate for patients with advanced disease is less than 30% (6). The poor survival rate of OC is partly attributable to the lack of effective prognostic biomarkers for individual patient. The existing prognostic factors for OC, such as radiological features (7), CA125 blood levels, histological features, and clinical stage (8), are insufficient for predicting individual clinical outcomes. Therefore, novel prognostic factors are urgently required for the early prediction of treatment outcomes to reduce the mortality rate of OC.
Genetic factors, such as gene expression alterations and gene mutations, have been considered to play a critical role in the regulation of OC development (9). Previous gene analysis studies have reported that genetic aberrations are involved in the pathogenesis of OC (10,11). For instance, Miles et al. found that RAD51AP1 was upregulated in OC (10), and Lee et al. reported that PIK3CA amplification caused cisplatin resistance in OC cell lines (12). Nevertheless, biomarkers with prognostic and predictive value still limit. Therefore, to identify an independent prognostic marker using genomic technologies and bioinformatics methods is urgently needed.
In the present study, we used a comprehensive bioinformatics analysis to probe the mechanisms and molecular markers associated with the prognosis of OC. Differentially expressed genes (DEGs) were screened out from 3 OC datasets (GSE26712, GSE18520, and GSE14407) from the Gene Expression Omnibus (GEO) database. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway functional enrichment analyses were performed by Metascape. The protein-protein interaction (PPI) network of the DEGs was constructed using the STRING database, and the prognostic values of hub genes were determined with the Kaplan-Meier (KM) plotter analysis. The ONCOMINE and Human Protein Atlas databases were used to identify the expression levels of prognostic genes in OC. The cBioPortal was used to find the mutations and amplification of prognostic genes. We also performed the function analyse for the prognostic genes.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-21-380).
Methods
Dataset integration and DEG screening
Three datasets (GSE26712, GSE18520, and GSE14407) were obtained from GEO for analysis of DEGs in OC. The files of the platform and the series of matrix files were downloaded, then calibrated and log-transformed using the R version 3.2.0. DEGs in the 3 microarray datasets above were screened out with the limma package (log FC >1 and adjusted P<0.05), and common DEGs among the datasets were identified using RobustRankAggreg. Package (P<0.05) and the heatmap of these DEGs was generated by heatmap package.
Functional enrichment analysis
To investigate the gene functions of the DEGs obtained from the 3 GEO datasets, Metascape (http://metascape.org) (13) was used to perform GO function and KEGG pathway enrichment analyses, with P<0.01 set as the cutoff for significance. To investigate the function of modules, the DAVID bioinformatics resource was also used.
PPI network construction
The PPI network of the DEGs was constructed using the STRING database (14) and Cytoscape v 3.6.0. The combined score of PPI was 0.468, the PPI enrichment P value was <0.01. Molecular Complex Detection (MCODE) clustering analysis was applied to find clusters of genes in the PPI network. The cutoff parameters were: degree cutoff =2; node score cutoff =0.2; k-core =2; and max. depth =100.
ONCOMINE analysis
The ONCOMINE microarray database (http://www.oncomine.org) is a website containing data on gene expressions in multiple cancers (15). We used ONCOMINE to compare the gene expression level of structural maintenance of chromosomes protein 4 (SMC4) in OC tissues with that in normal tissues. The screening conditions were set to log FC >2; P value <0.01; and top 10% gene rank.
KM plotter analysis
The prognostic values of the DEGs in OC were determined using the KM survival plotter (www.kmplot.com), an online database containing gene expression profiles and clinical data from public databases (16). The hub genes in the PPI network were input into the database to examine their associations with the survival prognosis and of patients with OC. KM survival plots were used to compare the overall survival (OS) and progression-free survival (PFS) of the low and high expression groups. The threshold for follow-up was 120 months.
Expression of SMC4 in OC tissues
The Human Protein Atlas v19 (http://www.proteinatlas.org/) is a database containing huge numbers of histological images of multiple tumors (17). We selected immunohistochemical (IHC) staining images from the database to compare the protein expression of SMC4 between OC tissues and normal tissues.
Mutations of SMC4 gene in OC
The cBio Cancer Genomics Portal (cBioPortal) database (http://cbioportal.org) explores and visualizes a large amount of genomic data of patients with cancer from sources including the Cancer Genome Atlas (TCGA) (18). We used cBioPortal to analyze somatic mutations and DNA copy number alterations (CNAs) of SMC4 in OC. We further analyzed the genetic alterations of SMC4 using data from UCSC Xena browserr (http://xena.uscs.edu/) (19), and copy number data were categorized into 4 groups as follows: shallow deletions, diploids, gains, and amplifications. UALCAN (http://ualcan.path.uab.edu) is a comprehensive interactive web tool based on level 3 RNA sequencing and clinical data of 31 cancer types in the TCGA database (20). We used UALCAN to identify the genes positively correlated with SMC4 and performed a functional pathway analysis of these genes using the DAVID bioinformatics resource.
GSEA
The Gene set enrichment analysis (GSEA) v3.0 software (www.broadinstitute.org/gsea/) was used to perform the functional pathway analysis of the SMC4 gene (21). Expression data were classified into high and low expression subgroups. Then, the enrichment analysis was done by employing the default weighted enrichment statistical method. The random combinatorial count was set to 1,000 times.
Statistical analysis
Several packages in R software were used to finish statistical calculations and graphs. The Kaplan-Meier plotter was employed to generate survival curves, and the threshold for follow-up was 120 months. P<0.05 and logFC ≥1 were considered as statistically significant for screening DEGs. log FC >2 and P<0.01 was set as statistically significant for the ONCOMINE analysis. P<0.01 was set to construct PPI network and perform enrichment analysis.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of DEGs in the GEO database
From the GSE14407 dataset, 2,756 DEGs (1,137 upregulated genes and 1,619 downregulated genes) were screened out. From the GSE18520 dataset, 2,623 DEGs (1,037 upregulated genes and 1,586 downregulated genes) were identified. From the GSE26712 dataset, 1,200 DEGs (444 upregulated genes and 756 downregulated genes) were screened out. The common DEGs identified from the 3 datasets were analyzed with RobustRankAggreg, and finally, a total of 879 common DEGs were obtained. The top 20 upregulated DEGs and downregulated DEGs are shown in a heatmap in Figure 1.
Enrichment analysis of DEGs
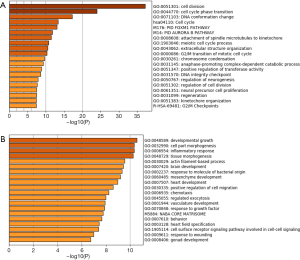
Metascape was used to perform function enrichment analyses and the top 20 Go and KEGG pathways were plotted. For GO term enrichment analysis, the results showed that the upregulated DEGs were predominantly enriched in cell division, cell cycle phase transition, DNA conformation change. KEGG analysis revealed that the upregulated DEGs were mostly enriched in cell cycle, G2/M checkpoints (Figure 2A). For GO term enrichment analysis, the downregulated DEGs were significantly enriched in developmental growth, cell part morphogenesis, inflammatory response (Figure 2B).
PPI network
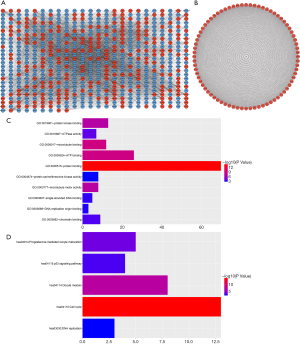
The PPI network had 626 nodes and 6,353 interactions (Figure 3A). Further, an important module containing 76 nodes and 2,656 interactions was identified from the PPI network (Figure 3B). The top 10 hub genes showing significant interactions were ZWINT, SMC4, NDC80, NEK2, AURKB, CENPF, KIF20A, KIF11, FAM83D, and CENPE. The GO analyses showed that the genes in the module were mainly enriched in protein binding, ATP binding (Figure 3C). The KEGG pathway analysis showed that these genes were enriched in cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation and DNA replication (Figure 3D).
Expression of SMC4 in common tumor types
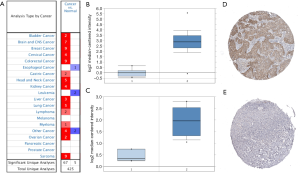
Using ONCOMINE, we found 425 studies that had investigated the mRNA level of SMC4 in human cancers compared with normal tissues. Of them, 67 studies reported that the expression levels of SMC4 were increased in human cancers including breast cancer, colorectal cancer, cervical cancer and OC, while 5 studies reported the downregulation of SMC4 (Figure 4A). Studies showed that SMC4 was highly expressed in OC tissues compared to normal tissues (Figure 4B,C). Subsequently, we validated the protein expression levels of SMC4 by performing an analysis using the Human Protein Atlas. The results showed SMC4 to be upregulated in OC tissues and downregulated in normal tissues (Figure 4D,E).
Genetic alterations of SMC4 in OC
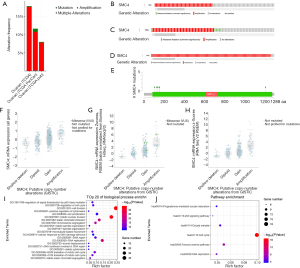
We futher discussed the causes of SMC4 overexpression in OC. From cBioPortal database, SMC4 alterations accounted for 7 to 18% of genetic alterations in OC. And alterations included somatic mutations, amplifications, and multiple alterations (Figure 5A,B,C,D). Notably, we found that the majority of SMC4 alterations were copy number amplifications. Missense mutation and embedded deletion (Figure 5C,E) were also frequently detected. We also analyzed the genetic alterations of SMC4 using the UCSC Xena browser. The copy number data in the TCGA were download and categorized into 4 groups (shallow deletions, diploids, gains, and amplifications). With the amplification of copy numbers, the corresponding SMC4 gene expression increased significantly (Figure 5F,G,H). We found that SMC4 copy number amplification was related to high SMC4 expression in OC. UALCAN, a tool for in-depth analysis of TCGA data, was utilized to verify the correlated hub genes of SMC4. Then, GO and KEGG pathway analyses of SMC4 and its related hub genes were performed. The GO analysis showed that those genes were significantly enriched in regulation of signal transduction by P53 class mediator, regulation cell cycle, cell division and so on. The KEGG pathway analysis showed that the genes were enriched in progesterone-mediated oocyte maturation, the P53 signaling pathway, oocyte meiosis, cell cycle and DNA replication (Figure 5I,J).
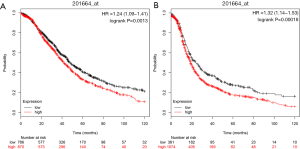
KM Plotter analysis
As shown in Figure 6, the prognostic value of SMC4 expression in OC was also analyzed. A high expression of SMC4 was correlated with significantly shorter OS and PFS in patients with OC (Figure 6A,B). Therefore, the SMC4 was subsequently screened as a putative prognostic marker for OC.
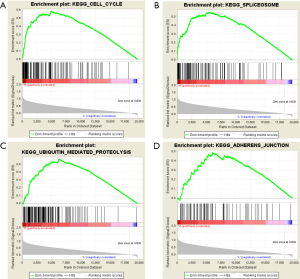
Enrichment analysis of SMC4 gene
SMC4 was found to be highly enriched in the cell cycle, spliceosome, ubiquitin mediated proteolysis, and adherens junctions (Figure 7A,B,C,D), which suggested that SMC4 might promote the development of OC via these signaling pathways.
Discussion
Novel biomarkers for OC could inform personalized treatment decisions and aid in the early prediction of prognosis for patients who are at high risk of disease recurrence and death. Unfortunately, prognostic biomarkers for OC are lacking. Here, we integrated gene expression data from the GEO datasets GSE26712, GSE18520, and GSE14407, and utilized muti-bioinformatics methods to explore potential prognostic biomarkers for OC. We identified 879 common DEGs from the 3 gene expression datasets. By constructing the PPI network, we screened out 10 hub genes in significant modules. The roles of the hub genes in the progression of OC were confirmed by the results of functional analyses. Furthermore, we found that 1 hub gene in the PPI network, SMC4, had a high expression at both the mRNA and protein levels, and an increased expression of SMC4 was associated with an unfavorable prognosis of OC. The results of analysis using cBioPortal showed that SMC4 alterations accounted for 7% to 18% of genetic alterations in OC. The majority of SMC4 alterations were copy number amplifications. Increased copy numbers levels were correlated with higher SMC4 expression. UALCAN was utilized to verify the hub genes co-expressed with SMC4. Finally, GSEA analysis showed that SMC4 were mainly enriched in the cell cycle, spliceosome, ubiquitin mediated proteolysis, and adherens junctions. SMC4 was subsequently identified as a prognostic biomarker closely associated with biological function, which might facilitate the prediction of prognosis in patients with OC.
SMC4 belongs to the structural maintenance of chromosomes (SMC) family, the members of which are involved in many physiological and pathological processes (22,23). SMC4 is highly expressed in multiple tumors, including hepatocellular carcinoma (24), colorectal cancer (25), prostate cancer (26), and lung cancer (27). Its high expression in hepatocellular carcinoma is responsible for tumor dedifferentiation and vascular invasion, and is correlated to an advanced disease stage (28). In Zhao et al.’s study, knockdown of SMC4 via RNA interference reduced prostate cancer cell migration and invasion (29). In another study, SMC4 overexpression was found to promote the proliferative, migratory, invasive capabilities of glioma cells (30). The above studies suggested that SMC4 played an important role in multiple tumors; however, its expression and function in OC were poorly understood. In the present study, we discovered abnormal expression and amplification of the SMC4 in OC. Our observations indicated that over expression SMC4 contributed to the tumorigenicity and prognosis of OC.
We also hypothesized that the aberrant expression of SMC4 affected the outcomes of patients with OC by regulating various signaling pathways. We found that SMC4 activates signaling pathways related to the cell cycle, spliceosome, ubiquitin mediated proteolysis, and adherens junctions, which were involved in the development of OC.
Dysregulation of the cell cycle pathway is important to tumorigenesis (31-34). SMC4 is associated with dysregulation of the cell cycle. For instance, Jiang et al. found that SMC4 upregulation drastically increased the proliferative capability of glioma cells by accelerating G1-S-phase transition (30). In A549 cells, knockdown of SMC4 was also observed to downregulate the cell cycle-related proteins cyclin B1 and cyclin-dependent kinase 1 to suppress cell proliferation (27). Thus, we postulate that SMC4 affected cell proliferation in OC via cell cycle regulation.
Our study also found that upregulation of SMC4 plays an essential role in OC via spliceosome, ubiquitin mediated proteolysis and adherens junctions. Aberrations of the spliceosome pathway have been shown to be involved in multiple processes in cancer, such as invasion, metastasis, and angiogenesis (35,36). Spliceosomal pathway genes affected the proliferation and metastasis of hepatocellular carcinoma (37). Ubiquitin mediated proteolysis has emerged in multiple cancers, and it was responsible for oncogenic transformation for glioma (38). Adherens junction controled normal cell-cell adhesion, might play an vital role in driving cancer cell dissemination (39). However, these pathway has rarely been reported in OC. Further investigation of the relationship between the regulation of these pathways and development of OC should be also carried out.
Our preliminary study has confirmed SMC4 as a prognostic biomarker in OC and its’ activated signaling pathways. However our study had some limitations, the experiments in vitro and in vivo for validation should be conducted. We would further discuss the specific impacts of SMC4 on resistance of platinum-based chemotherapy and recurrence in OC, which are main reasons for treatment failure and main factor affecting outcome in future study.
Conclusions
In the present study, we used a comprehensive bioinformatics analysis to identify DEGs in patients with OC. We finally identified SMC4 to have vital involvement in OC, and its overexpression to be closely associated with a poor prognosis in patients with the disease. We also found that SMC4 plays important roles in the biological processes of OC. As a conclusion, we could tell that SMC4 is a useful and novel prognostic indicator for patients with OC. Further molecular biological experiments should be performed to illuminate the biological characteristics of SMC4 in OC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-21-380
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-21-380). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2018: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 2018;68:297-316. [Crossref] [PubMed]
- Goodman MT, Howe HL. Descriptive epidemiology of ovarian cancer in the United States, 1992-1997. Cancer 2003;97:2615-30. [Crossref] [PubMed]
- Dembo AJ, Davy M, Stenwig AE, et al. Prognostic factors in patients with stage I epithelial ovarian cancer. Obstet Gynecol 1990;75:263-73. [PubMed]
- Kinose Y, Sawada K, Nakamura K, et al. The role of microRNAs in ovarian cancer. Biomed Res Int 2014;2014:249393. [Crossref] [PubMed]
- Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol 2000;19:3-10. [Crossref] [PubMed]
- Annual report on the results of treatment in gynecological cancer. Twenty-first volume. Statements of results obtained in patients treated in 1982 to 1986, inclusive 3 and 5-year survival up to 1990. Int J Gynaecol Obstet 1991;36:1-315. [PubMed]
- Prayer L, Kainz C, Kramer J, et al. CT and MR accuracy in the detection of tumor recurrence in patients treated for ovarian cancer. J Comput Assist Tomogr 1993;17:626-32. [Crossref] [PubMed]
- Tingulstad S, Skjeldestad FE, Halvorsen TB, et al. Survival and prognostic factors in patients with ovarian cancer. Obstet Gynecol 2003;101:885-91. [PubMed]
- Baylin SB, Esteller M, Rountree MR, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 2001;10:687-92. [Crossref] [PubMed]
- Miles GD, Seiler M, Rodriguez L, et al. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes 2012;5:164. [Crossref] [PubMed]
- Bartlett JM, Langdon SP, Simpson BJ, et al. The prognostic value of epidermal growth factor receptor mRNA expression in primary ovarian cancer. Br J Cancer 1996;73:301-6. [Crossref] [PubMed]
- Lee S, Choi EJ, Jin C, et al. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol 2005;97:26-34. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362-D368. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas-a tool for pathology. J Pathol 2008;216:387-93. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Tyner C, Barber GP, Casper J, et al. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 2017;45:D626-34. [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 2012;26:1659-78. [Crossref] [PubMed]
- Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem 1999;263:6-13. [Crossref] [PubMed]
- Zhou B, Chen H, Wei D, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res 2014;33:55. [Crossref] [PubMed]
- Jinushi T, Shibayama Y, Kinoshita I, et al. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med 2014;3:1544-52. [Crossref] [PubMed]
- Haram KM, Peltier HJ, Lu B, et al. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate 2008;68:1517-30. [Crossref] [PubMed]
- Zhang C, Kuang M, Li M, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci Rep 2016;6:34508. [Crossref] [PubMed]
- Zhou B, Yuan T, Liu M, et al. Overexpression of the structural maintenance of chromosome 4 protein is associated with tumor de-differentiation, advanced stage and vascular invasion of primary liver cancer. Oncol Rep 2012;28:1263-8. [Crossref] [PubMed]
- Zhao SG, Evans JR, Kothari V, et al. The Landscape of Prognostic Outlier Genes in High-Risk Prostate Cancer. Clin Cancer Res 2016;22:1777-86. [Crossref] [PubMed]
- Jiang L, Zhou J, Zhong D, et al. Overexpression of SMC4 activates TGFβ/Smad signaling and promotes aggressive phenotype in glioma cells. Oncogenesis 2017;6:e301. [Crossref] [PubMed]
- Masciullo V, Scambia G, Marone M, et al. Altered expression of cyclin D1 and CDK4 genes in ovarian carcinomas. Int J Cancer 1997;74:390-5. [Crossref] [PubMed]
- Foster I. Cancer: A cell cycle defect. Radiography 2008;14:144-9. [Crossref]
- Maddika S, Ande SR, Panigrahi S, et al. Cell survival, cell death and cell cycle pathways are interconnected: implications for cancer therapy. Drug Resist Updat 2007;10:13-29. [Crossref] [PubMed]
- Coletta RD, Jedlicka P, Gutierrez-Hartmann A, et al. Transcriptional control of the cell cycle in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia 2004;9:39-53. [Crossref] [PubMed]
- Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol 2007;39:1432-49. [Crossref] [PubMed]
- van Alphen RJ, Wiemer EA, Burger H, et al. The spliceosome as target for anticancer treatment. Br J Cancer 2009;100:228-32. [Crossref] [PubMed]
- Hou G, Liu G, Yang Y, et al. Neuraminidase 1 (NEU1) promotes proliferation and migration as a diagnostic and prognostic biomarker of hepatocellular carcinoma. Oncotarget 2016;7:64957-66. [Crossref] [PubMed]
- Liang R, Wang M, Zheng G, et al. A comprehensive analysis of prognosis prediction models based on pathwaylevel, genelevel and clinical information for glioblastoma. Int J Mol Med 2018;42:1837-46. [PubMed]
- Gloushankova NA, Rubtsova SN, Zhitnyak IY. Cadherin-mediated cell-cell interactions in normal and cancer cells. Tissue Barriers 2017;5:e1356900. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)



