
Integrative bioinformatics analysis identifies LINC01614 as a potential prognostic signature in esophageal cancer
Introduction
Esophageal cancer (EC), one of the most common cancers, is ranking eighth in the incidence of all the cancer types and sixth in the causable factors in cancer-related death, and leads a malignancy causing lots of mortality worldwide (1,2). Esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC) are known as the two main histological types of EC (2). ESCC is the major type of EC which predominates in the Asian countries, whereas EAC is more predominant throughout the Western countries (3).
As far as we know, the development of EC is a complex process involving multistep and multiple factors. Recently, long non-coding RNAs (lncRNAs) as a novel biomarker in human cancer is gradually being recognized. LncRNAs are longer than 200 nucleotides with less or no protein coding capacity and widely involved in the gene regulations at different levels (epigenetic regulations, transcriptional, and posttranscriptional processing) (3-6). The well-studied functions of lncRNAs include sequence-specific recruitment of proteins, competing endogenous RNA (ceRNA) regulation and molecular scaffolding of protein complexes (7). Abnormal content or functions of lncRNAs have been demonstrated to be associated with tumorigenesis and metastasis, suggesting that lncRNAs may be novel biomarkers for tumor diagnosis and potential therapeutic targets (8). Although many functional lncRNAs have been identified using bioinformatics platform and various cellular functional experiments in EC (9-11), a large number of lncRNAs related to EC progression remain to be further studied, which may provide new strategy for further development of tumor targeted drug and early diagnosis in patients with EC. Thus, analyzing the clinical significance, biological function and molecular mechanisms of lncRNAs in EC is of great importance.
The gene encoding long intergenic non-protein-coding RNA 1614 (LINC01614), located on chromosome 2q35 in between two exons, is known as lung cancer-associated lncRNA 4 and was also originally identified as a tumor promoter in lung cancer (12-14). Then, LINC01614 was also reported to be overexpressed in breast cancer (15), glioma (16), and was proved to have great prognostic values in cancer patients. Through the LINC01614-contaitng signature predicted overall survival (OS) and disease free survival (DFS) in patients with ESCC (17), the expression and biological function of LINC01614 in EC are unclear.
Integrated analysis of accumulated data is an effective way to obtain new biomarkers or potential therapeutic targets for EC. So, the datasets belonging to GPL18109 (a platform that specifically applied to identify lncRNAs and mRNAs) and TCGA were downloaded for DE lncRNAs and DE mRNAs analysis in this study. And the online tool GEPIA2 was used to analyze prognostic values for lncRNAs and their co-expression mRNAs in EC. Next, we used bioinformatics tools to construct a lncRNA-mRNAs network in EC. Furthermore, we found that LINC01614 served as an oncogene that could promote cell proliferation and migration in EC. Overall, our study showed a novel potentially clinical lncRNA biomarker for diagnosis and prognosis in EC.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2529).
Methods
Public data collection and bioinformation analyses
The details of three datasets (GSE53622, GSE53624, and GSE53625) were downloaded from GEO database, and EC RNA expression profiles were downloaded from TCGA database. We performed differentially expressed (DE) mRNAs and lncRNAs with thresholds of |log fold change| >1 and adjusted P value <0.05 by comparing tumor tissues with normal tissues using limma R package. The integrated dysregulated gene lists were saved for subsequent analysis. An online tool GEPIA2 was used to analyze the hub lncRNAs and mRNAs of EC specimens. Our workflow for bioinformatics analysis is illustrated in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and siRNA transfection
Human EC cell lines (EC9706, TE-1, and EC109) and human normal esophageal epithelial cells (HEEC) were purchased from the Chinese Academy of Cell Resource Center (Shanghai, China). Cell lines were cultured in RPMI-1640 (Corning) supplemented with 10% Fetal Bovine Serum (FBS) (Invitrogen), 100 μg/mL streptomycin, and 100 IU/mL penicillin. All cells were grown at 37 °C in a humidified incubator with 5% CO2. The small interfering RNAs (siRNAs) targeting LINC01614 (si-LINC01614#1, si-LINC01614#2) and negative control siRNAs (si-NC) were synthesized by GenePharma and used in knockdown experiments. 50 nM target siRNAs were transfected into EC109 and TE-1 cells using Lipofectamine 3000 (Invitrogen). The sequences of siRNAs are shown in Table S1.
RNA isolation and RT-PCR
Following the manufacturer’s instruction, total RNAs were isolated using a Trizol reagent (TIANGEN, Beijing, China). As previously described, the Reverse Transcription-Polymerase Chain Reaction (RT-PCR) were performed with Fastking RT Reagent Kit (TIANGEN, Beijing, China) and SYB Green Premix kit (TIANGEN, Beijing, China). The results of RT-PCR were analyzed by the 2−ΔΔCT formula. The primers used in this study were shown in Table S2, and GAPDH was selected as an internal reference.
Cell viability assay
After transfection, EC109 and TE-1 cells were seeded into 96-well plate at 2,000 cells per well with 100 μL complete medium. CCK8 regent (10 μL/well, Solarbio) was incubated with cells for 2.5 h at 37 °C and the OD450 was measured by microplate reader (Tecan) per day up to 5 days.
Transwell migration assay
Migration assay was performed using Transwell chambers (Corning, 3422). After transfection, EC109 (6×104 cells/well/100 μL) or TE-1 (8×104 cells/well/100 μL) cells were seeded onto the upper chambers with serum-free RPMI 1640, and 600 μL RPMI 1640 with 30% FBS medium were filled in the lower chamber and incubated at 37 °C for 24 h. Cells in lower chamber were fixed with 1% paraformaldehyde for 20 min, and stained with Giemsa. Images of cells were taken and analyzed using National Institutes of Health (NIH) imageJ software.
Flow cytometry
siLINC01614#1/2 or siNC was transfected into EC109 and TE-1 cell lines respectively. The effects of LINC01614 on cell apoptosis were analyzed by flow cytometry using the annexin V-APC/PI apoptosis kit (Beyotime, China) according to protocol. PI can distinguish live cells from dead cells with the use of a counterstaining dye.
Statistical analysis
The statistical results came from three independent experiments. Results were analyzed using SPSS 26 and Prism 7.0 (GraphPad). Student’s t-test was used to analyze the statistical significance and multiple groups were compared by one-way ANOVA. Data are expressed as the mean ± SD. A value of P<0.05 was considered as statistical significance.
Results
Differentially expressed lncRNAs and mRNAs
In our study, three datasets supplemented with their platform information were downloaded from GEO database, and EC RNA expression profiles were downloaded from TCGA database. Before integrated analysis of these datasets, limma package was applied to screen differentially expressed genes (DEGs) according to the threshold of |Log Fold change| >1 and adjusted P value <0.05. The detailed information of the three GEO datasets were shown in Table S3. We found 1033 DE lncRNAs (663 upregulated and 370 downregulated) and 3,562 DE mRNAs (1,925 upregulated and 1,637 downregulated) in ESCA-TCGA dataset. The intersection of the DE genes with consistent RNA levels from GEO and TCGA datasets includes 24 DE lncRNAs (18 upregulated and 6 downregulated) and 659 DEmRNAs (379 upregulated and 280 downregulated).
Identification of a prognostic signature of EC
The clinical information of EC patients in the TCGA database was shown in Table S4. To validate the association between lncRNAs and EC, we first selected 20 DE lncRNAs (4 DE lncRNAs reported in EC was out) for OS and DFS analysis using GEPIA2. And we found that only upregulated LINC01614 was associated with short recurrence-free survival (RFS) in EC (Figure 2A). Moreover, the correlation between the expression of Linc01614 and the clinical pathology of EC revealed that LINC01614 was associated with American Journal of Critical Care (AJCC) stage and T stage (Figure 2B,C and Table S5). RT-PCR results showed that the expression levels of LINC01614 were higher in EC tissues than in non-tumor tissues (Figure 2D).These results indicate that LINC01614 may play an important role in EC development.
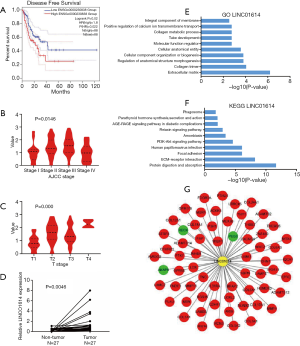
Functional enrichment analysis and Construction of lncRNA-mRNAs network
To further investigate the function of LINC01614, we firstly obtained 1,000 co-expression genes of LINC01614 from GEPIA2 in EC. Secondly, we selected 64 mRNAs (Table S6), which came from the intersection of GEPIA2 and DE mRNAs from TCGA and GEO, for functional enrichment analysis using KOBAS. The top 10 biological processes identified in Gene Ontology (GO) and KEGG pathways revealed that mRNAs positively co-expressed with LINC01614 participated primarily in “extracellular matrix”, “cellular component organization or biogenesis”, “ECM-receptor interaction”, “Focal adhesion” and “PI3K-Akt signaling pathway” (Figure 2E,F), suggesting that LINC01614 may regulate tumorigenesis in EC. Finally, we selected the 64 mRNAs for construction of LINC01614-mRNAs network (Figure 2G), which was visualized using Cytoscape v3.7.2.
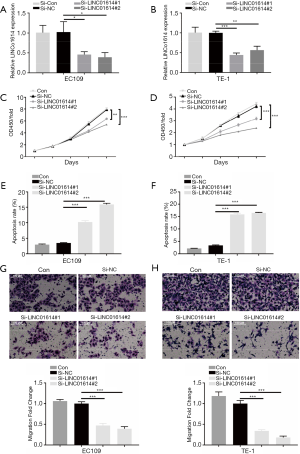
LINC01614 promotes EC cell proliferation and migration
To further verify the role of LINC01614 in EC cell lines, we knocked down its expression via transfecting siRNA targeted LINC01614 into EC109 or TE-1 cells. RT-PCR results showed that siRNAs were successful transduction of EC cells (Figure 3A,B). The following CCK8 assays and cell apoptosis assays confirmed that knockdown of LINC01614 inhibited cell proliferation and induced cell apoptosis in EC109 and TE-1 cells (Figure 3C,D,E,F). Moreover, Transwell assays showed that knockdown of LINC01614 significantly inhibited both EC109 and TE-1 cells migration, compared with control cells (Figure 3G,H). In conclusion, LINC01614 could promote EC cell proliferation and migration.
Discussion
EC is one of the most common gastrointestinal cancers and the incidence is on the increase in recent years (18,19). Due to lack of early diagnostic strategy, the patients with EC exhibits at a late stage in the present of diagnosis and their 5-year survival rate is extremely poor, only 15% to 25% (20). Increasing evidence have reported that non-coding RNAs (ncRNAs) and protein coding mRNA play a pivotal in many important biological processes, including cell proliferation, differentiation, gene expression, cell apoptosis, migration and invasion (8,21,22), especially in tumorigenesis. However, only a few lncRNAs have been functionally characterized in EC. Nowadays, several lncRNAs has been reported closely associated with the prognosis of EC patients, such as LINC01415 (23), LINC01980 (24,25), LINC02042 (26), and PVT1 (27). Interestingly, growing studies hold that the dysregulation of lncRNAs was caused by various transcription factors. For instance, SP1 increased lncRNA-ZFAS1 in colorectal cancer, resulting in the increase of the capacities of cell proliferation, migration and invasion (28). Hence, it might be very necessary for seeking new biomarkers and therapeutic targets of lncRNAs for EC.
In our study, we firstly identified the DE lncRNAs involved in EC by analyzing gene expression data obtained from GEO and TCGA databases. Using online bioinformatics tools, we found that LINC01614 was associated with short RFS, advanced AJCC stage and T stage in EC, which indicated that LINC01614 might be a novel biomarker of EC. A clinical study indicated that high LINC01614 was closely associated with advanced AJCC stage and shorter DFS of EC patients and could serve as an independent prognostic factor for EC patients. Furthermore, our findings revealed that 64 mRNAs were observed to consist for the construction of LINC01614-mRNAs network in EC. To further investigate the role of LINC01614, we build the interaction network for 64 mRNAs and LINC01614 from GO and KEGG analysis. Using these data, we found LINC01614 are involved in different terms of biological process related to tumorigenesis. The GO terms for up-regulated LINC01614 are extracellular matrix, and cellular component organization or biogenesis. Meanwhile, we found the most significant KEGG pathways are ECM-receptor interaction and PI3K-Akt signaling pathway, which have been reported to be associated with the progression of EC. Previously, the oncogene roles of LINC01614 in glioma, breast cancer and lung adenocarcinoma has been reported recently (13,15,16). Furthermore, we confirmed the silico analysis results by lost-of-function assays. CCK8, cell apoptosis and Transwell assays showed that knockdown of LINC01614 inhibited EC cell proliferation and migration. Besides, LINC01614 may protect EC cells from apoptosis. Finally, we confirmed LINC01614 as a novel biomarker and drove tumorigenesis via promoting tumor growth and metastasis in EC.
Therefore, from data in the present study, we learn that LINC01614 might affect tumorigenesis through regulating protein-coding mRNAs, which were involved in cell development, proliferation, ECM formation, PI3K-Akt signaling pathway and other important biological processes. However, there are still several questions to be addressed as follows: Does LINC01614 bind to DE mRNAs directly or indirectly? How does LINC01614 identify and interact with precise genomic target sites? What are the specific mechanisms of LINC01614 in vivo? We will focus on these issues in our future work.
In conclusion, we verified LINC01614 as an oncogene in EC by bioinformatics and CCK8, Transwell and flow cytometry experiments. And co-expression network for all of these DE mRNAs and LINC01614 were built to explore the relationship between mRNA and lncRNA, which may provide a mechanism of mRNA expression regulation by lncRNA. Our study provide insight into the identification of novel lncRNA biomarkers for diagnosis and prognosis in EC.
Acknowledgments
We thank Lareina Zhao for her linguistic assistance during the preparation of this manuscript.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2529
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2529). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Q, Dai Z, Xia C, et al. Suppression of long non-coding RNA MALAT1 inhibits survival and metastasis of esophagus cancer cells by sponging miR-1-3p/CORO1C/TPM3 axis. Mol Cell Biochem 2020;470:165-74. [Crossref] [PubMed]
- Zhang XM, Wang J, Liu ZL, et al. LINC00657/miR-26a-5p/CKS2 ceRNA network promotes the growth of esophageal cancer cells via the MDM2/p53/Bcl2/Bax pathway. Biosci Rep 2020;40:BSR20200525. [Crossref] [PubMed]
- Wu Y, Yang X, Chen Z, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 2019;18:87. [Crossref] [PubMed]
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta 2014;1839:1097-109. [Crossref] [PubMed]
- Suzuki H, Maruyama R, Yamamoto E, et al. Relationship Between Noncoding RNA Dysregulation and Epigenetic Mechanisms in Cancer. Adv Exp Med Biol 2016;927:109-35. [Crossref] [PubMed]
- Wu Z, Liu X, Liu L, et al. Regulation of lncRNA expression. Cell Mol Biol Lett 2014;19:561-75. [Crossref] [PubMed]
- Zhang X, Jiang Y, Xie Y, et al. Comprehensive Analysis of lncRNAs Associated with the Pathogenesis and Prognosis of Gastric Cancer. DNA Cell Biol 2020;39:299-309. [Crossref] [PubMed]
- Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Chen MJ, Deng J, Chen C, et al. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol 2019;113:27-36. [Crossref] [PubMed]
- Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ 2018;25:1980-95. [Crossref] [PubMed]
- Liu H, Zhang Q, Lou Q, et al. Differential Analysis of lncRNA, miRNA and mRNA Expression Profiles and the Prognostic Value of lncRNA in Esophageal Cancer. Pathol Oncol Res 2020;26:1029-39. [Crossref] [PubMed]
- White NM, Cabanski CR, Silva-Fisher JM, et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 2014;15:429. [Crossref] [PubMed]
- Liu AN, Qu HJ, Yu CY, et al. Knockdown of LINC01614 inhibits lung adenocarcinoma cell progression by up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med 2018;22:4034-44. [Crossref] [PubMed]
- Sun Y, Ling C. Analysis of the long non-coding RNA LINC01614 in non-small cell lung cancer. Medicine (Baltimore) 2019;98:e16437. [Crossref] [PubMed]
- Vishnubalaji R, Shaath H, Elkord E, et al. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov 2019;5:109. [Crossref] [PubMed]
- Wang H, Wu J, Guo W. SP1-Mediated Upregulation of lncRNA LINC01614 Functions a ceRNA for miR-383 to Facilitate Glioma Progression Through Regulation of ADAM12. Onco Targets Ther 2020;13:4305-18. [Crossref] [PubMed]
- Huang GW, Xue YJ, Wu ZY, et al. A three-lncRNA signature predicts overall survival and disease-free survival in patients with esophageal squamous cell carcinoma. BMC Cancer 2018;18:147. [Crossref] [PubMed]
- Hou X, Wen J, Ren Z, et al. Non-coding RNAs: new biomarkers and therapeutic targets for esophageal cancer. Oncotarget 2017;8:43571-8. [Crossref] [PubMed]
- Pakzad R, Mohammadian-Hafshejani A, Khosravi B, et al. The incidence and mortality of esophageal cancer and their relationship to development in Asia. Ann Transl Med 2016;4:29. [PubMed]
- Ohashi S. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology 2015;149:1700-15. [Crossref] [PubMed]
- Yu M, Yu S, Gong W, et al. Knockdown of linc01023 restrains glioma proliferation, migration and invasion by regulating IGF-1R/AKT pathway. J Cancer 2019;10:2961-8. [Crossref] [PubMed]
- Tan D, Wu Y, Hu L, et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus 2017;30:1-9. [PubMed]
- Shi X, Li Y, Sun Y, et al. Genome-wide analysis of lncRNAs, miRNAs, and mRNAs forming a prognostic scoring system in esophageal squamous cell carcinoma. PeerJ 2020;8:e8368. [Crossref] [PubMed]
- Zhang S, Liang Y, Wu Y, et al. Upregulation of a novel lncRNA LINC01980 promotes tumor growth of esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2019;513:73-80. [Crossref] [PubMed]
- Liang X, Wu Z, Shen S, et al. LINC01980 facilitates esophageal squamous cell carcinoma progression via regulation of miR-190a-5p/MYO5A pathway. Arch Biochem Biophys 2020;686:108371. [Crossref] [PubMed]
- Du J, Zhang G, Qiu H, et al. A novel positive feedback loop of linc02042 and c-Myc mediated by YBX1 promotes tumorigenesis and metastasis in esophageal squamous cell carcinoma. Cancer Cell Int 2020;20:75. [Crossref] [PubMed]
- Shen SN, Li K, Liu Y, et al. Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol Oncol 2019;13:2554-73. [Crossref] [PubMed]
- Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis 2018;9:982. [Crossref] [PubMed]



