
Prognostic value and potential molecular mechanism of the like-Sm gene family in early-stage pancreatic ductal adenocarcinoma
Introduction
About 90% of pancreatic cancers originate from the epithelium of pancreatic ducts, and pancreatic ductal adenocarcinoma (PDAC) is one of the most deadly pancreatic cancers (1). The mortality rate of PDAC is astonishingly high, with 333,000 new PDAC cases diagnosed worldwide every year, and although only accounting for 3.1% of all cancers, more than 300,000 new cases of PDAC die every year. Pancreatic cancer is now the fourth leading cause of cancer-related deaths in the United States, and the annual survival rate is only 7.7% (2,3). A total of 46,420 patients were diagnosed with PDAC in the United States in 2014; it is expected that by the end of 2020, the number of PDAC patients in the United States will double, and by 2030 PDAC is expected to be the second leading cause of cancer deaths worldwide, after lung cancer, indicating poor prognosis (1,4,5).
About 70% of PDAC deaths are caused by cancer metastasis (6), and this coupled with a lack of early warning symptoms and a highly malignant phenotype are the leading causes of high mortality in PDAC patients. Many patients are already at an advanced stage of cancer when diagnosed; thus, early detection and early treatment are crucial for defeating PDAC (1,6,7). Despite tremendous progress in recent years in surgery, radiotherapy, and chemotherapy, the prognosis for pancreatic cancer remains poor. Although systemic chemotherapy is an essential treatment for liver and peritoneal metastasis, it has little effect on advanced pancreatic cancer (8,9), and molecular targeted therapy has no significant impact on the survival of PDAC patients (5). Developing a new PDAC biomarker, verifying its role in the early detection of PDAC, and predicting the prognosis of PDAC patients are therefore urgent for enhancing survival (3).
There are 14 genes in the like-sm (LSM) gene family; in animals, LSMs generally exist in the form of heptameric complexes, including the LSM1-7 and LSM2-8 complexes, and they are closely related to splicing and cytoplasmic mRNA degradation (10). LSM genes were significantly overexpressed in pancreatic cancer compared with surrounding normal tissues, and they act as oncogenes promoting cancer transformation, proliferation, resistance, and metastasis (11). The mechanism by which LSM genes promote PDAC transformation remains unclear. Studies have shown that up-regulation of LSM-1 can promote the growth of pancreatic tumors in mice, while down-regulation of LSM-1 can have the opposite effect (12), and antisense RNA induction significantly inhibits the ability of tumor cells to form anchoring-independent colonies on soft agar (13).
Herein, we used the TCGA database and found that in patients expressing high levels of LSM1 and LSM4 in pancreatic cancer, overall survival time (OS) was significantly decreased. This could provide a new direction for the treatment of pancreatic cancer. LSM genes are not only related to pancreatic cancer, but also closely related to other cancers such as prostate cancer and breast cancer. Cox multivariate proportional risk regression analysis showed that LSM1 and LSM4 may be early pancreas predictors, and are the primary genes involved in the proliferation and metastasis of pancreatic cancer. However, at present, most studies on the LSM gene family have only explored the role of LSM1 in pancreatic tumor transformation and its possible mechanism. In the present work, we comprehensively examined the roles of all members of the LSM gene family in early-stage PDAC, as well as the potential molecular mechanisms. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3056).
Methods
Data mining and processing
PDAC patient data (therapeutic function and mRNA profile) were downloaded from the TCGA website (https:/cancergenome.nih.gov/, accessed April 20, 2017). Clinical features included gender, smoking history, histological grade, presence or absence of Whipple surgery and radiotherapy, etc. After downloading, the DESeq package in R (version 3.5.2; www.r-project.org) was used to normalize expression data. All data used in this study were downloaded from a public database, and no humans or animals were involved in this research. Hence, no approval from an ethics committee was required. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Bioinformatics analysis of LSM genes
Gene Ontology (GO) was used for functional annotation of LSM genes. Additionally, the Kyoto Encyclopedia of Genes & Genomes (KEGG) database was accessed to define related signaling pathways, using the DAVID program (https://david.ncifcrf.gov/home.jsp, version 6.8, accessed January 19, 2019) (14). The Corrplot package (version 3.6.1) was applied (www.r-project.org) to analyze the correlation coefficients of LSM genes. We also used a search tool to construct a protein-protein interaction (PPI) map by retrieving gene/protein interactions (https://string-db.org/, version 11, accessed 30 October 2019). All LSM genes were uploaded to the Homo sapiens database, and an interaction score greater than 0.6 was deemed significant. Finally, we used GeneMANIA to construct the gene-gene interaction (GGI) network (http://genemania.org/, accessed October 30, 2019) (15).
Survival analysis
To review the partnership between specific therapeutic variables and survival prognoses in patients with early PDAC, Kaplan-Meier log-rank tests were applied. The multivariate Cox proportional risk regression system integrates related factors to determine whether LSM genes are strongly correlated with patient OS. Survival analysis of a single gene, comprehensive effect survival analysis based on the results, clinical variables, and expression levels of LSM genes were analyzed in R. A nomogram was generated using an RMS kit. To evaluate the PDAC prognosis of LSM genes, the Kaplan-Meier log-rank test was employed, along with the multivariate Cox proportional probability regression model.
Prognostic signature construction
After performing survival analysis, we found that LSM1 and LSM4 were significantly correlated with the prognosis of PDAC. Based on this, a predictive model of gene expression levels was constructed using the following risk scoring formula: Risk score = expression of gene 1×β1+ expression of gene 2×β2+ ... expression of Gene n×βn (16,17), where βn is the coefficient of regression of the related gene Cox threat analysis. Depending on the median risk assessment, high-risk individuals and low-risk patients are categorized into different groups. The ROC survival package in R (18) was used to develop a time-dependent receiver operator characteristic curve (ROC) for survival assessment to evaluate the prognosis of high-risk to low-risk groups.
Gene set enrichment analysis (GSEA)
Whole-genome expression profile datasets were uploaded and corresponding grouping files expressed by LSM genes were subjected to GSEA (19). The molecular feature database MSigDB (20) was used to expand c2 and c5 databases to determine in which pathways LSM genes are enriched. The upper or lower parts of the phenotype-related sorted gene list comprises LSM genes on each chromosome. A sequence of genes was considered statistically meaningful if both false discovery score (FDR) <0.25 and P<0.05 criteria were met.
Statistical analysis
For statistical analysis, we used SPSS 22.0 (IBM) and R version 3.6.1 (https:/www.r-project.org/). LSM gene relative risk between high- and low-expression categories was calculated using hazard-ratios (HRs) and a 95% confidence interval (CI). When evaluating the association of LSM genes, P<0.05 was regarded as statistically significant, and the Pearson correlation coefficient was employed (21-23).
Results
Data acquisition
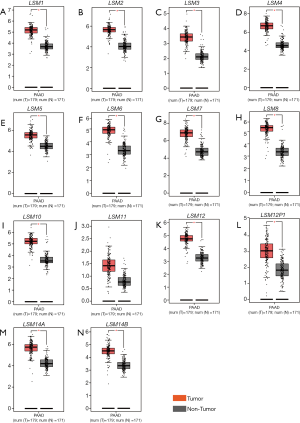
Clinical characteristics and transcription profiles of PDAC patients were obtained from the TCGA database. After excluding advanced-stage PDAC without Whipple surgery, data for 112 patients undergoing Whipple surgery were retained for further analysis. Gene information was entered into a gene expression profile (GEPIA, http://gepia.pku.cn/), default parameters were employed, and patients were divided into two groups based on the median expression values of genes. Expression of LSM1 and LSM4 in tumor tissues of patients with PDAC was significantly higher than in normal adjacent tissues (P<0.05), which indicates that LSM1 and LSM4 might be involved in tumorigenesis and development in PDAC (Figure 1).
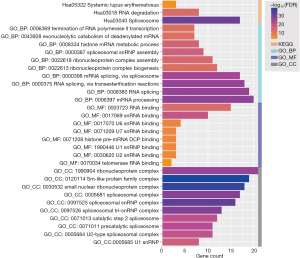
GO and KEGG enrichment analysis.
As shown in Figure 2, GO terms and KEGG pathway analysis revealed that the LSM gene family was mainly enriched in systemic lupus erythematosus, spliceosome, histone mRNA metabolic process, via transesterification reactions, histone mRNA metabolic process, mRNA binding, and ribonucleoprotein complex categories.
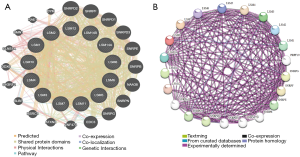
Analysis of LSM family interactions at the gene and protein levels
A complex gene-gene connection network was constructed between LSM family genes, and genes were clustered together (Figure 3A). Meanwhile, to explore the biological functions of LSM genes, we also performed PPI analysis to identify proteins that interact with LSMs. We identified SNRPD, SNRPF, EDC3, ATXN2, ZNF473, DDX6 and SLBP as potential interacting partners (Figure 3B). The Pearson’s correlation analysis matrix diagram (Figure 4) shows that LSM1, LSM2, and LSM5 are closely related (correlation coefficient >0.5). LSM2, LSM3, LSM5, and LSM7 are also closely related (correlation coefficient >0.5). LSM4 and LSM7 is the most closely related pair, with a correlation coefficient of 0.73. The number in each grid of the correlation graph represents the correlation between corresponding genes.
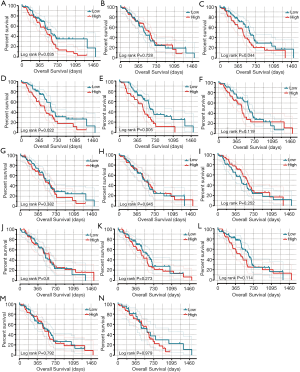
Survival analysis of the LSM gene family
As shown in Table S1, histological ratings, guided molecular therapy, radiation therapy, and residual resection are all substantially related to OS, based on a summary of clinical data for each patient. We found that high expression levels of LSM1 (adjusted P=0.004, adjusted HR =2.338, 95% CI: 1.320–4.141), and LSM4 (adjusted P=0.034, adjusted HR =1.803, 95% CI: 1.046–3.109) is associated with bad outcomes (Table 1 and Figure 5).
Table 1
| Gene expression | Events/total (n=112) | MST (days) | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI) | Adjusted P-value |
|---|---|---|---|---|---|---|
| LSM1 | ||||||
| Low | 30/56 | 593 | 1 | 1 | ||
| High | 39/56 | 498 | 1.681 (1.032–2.738) | 0.035 | 2.338 (1.320–4.141) | 0.004 |
| LSM2 | ||||||
| Low | 37/56 | 486 | 1 | 1 | ||
| High | 32/56 | 603 | 0.918 (0.568–1.484) | 0.728 | 0.707 (0.415–1.203) | 0.201 |
| LSM3 | ||||||
| Low | 30/56 | 614 | 1 | 1 | ||
| High | 39/56 | 473 | 1.638 (1.007–2.664) | 0.044 | 1.298 (0.746–2.258) | 0.356 |
| LSM4 | ||||||
| Low | 32/56 | 603 | 1 | 1 | ||
| High | 37/56 | 473 | 1.752 (1.078–2.846) | 0.022 | 1.803 (1.046–3.109) | 0.034 |
| LSM5 | ||||||
| Low | 31/56 | 614 | 1 | 1 | ||
| High | 38/56 | 473 | 1.997 (1.218–3.275) | 0.005 | 1.370 (0.750–2.501) | 0.306 |
| LSM6 | ||||||
| Low | 35/56 | 607 | 1 | 1 | ||
| High | 34/56 | 470 | 1.475 (0.903–2.411) | 0.119 | 0.951 (0.551–1.640) | 0.855 |
| LSM7 | ||||||
| Low | 36/56 | 518 | 1 | 1 | ||
| High | 33/56 | 511 | 1.239 (0.766–2.003) | 0.382 | 0.892 (0.520–1.529) | 0.677 |
| LSM8 | ||||||
| Low | 35/56 | 518 | 1 | 1 | ||
| High | 34/56 | 517 | 0.954 (0.592–1.536) | 0.845 | 0.800 (0.462–1.386) | 0.427 |
| LSM10 | ||||||
| Low | 38/56 | 476 | 1 | 1 | ||
| High | 31/56 | 592 | 0.757 (0.469–1.221) | 0.252 | 0.767 (0.459–1.283) | 0.313 |
| LSM11 | ||||||
| Low | 33/56 | 511 | 1 | 1 | ||
| High | 36/56 | 592 | 0.940 (0.583–1.517) | 0.8 | 0.954 (0.564–1.615) | 0.861 |
| LSM12 | ||||||
| Low | 28/56 | 607 | 1 | 1 | ||
| High | 41/56 | 481 | 0.762 (0.468–1.241) | 0.273 | 0.981 (0.575–1.674) | 0.943 |
| LSM12P1 | ||||||
| Low | 29/56 | 627 | 1 | 1 | ||
| High | 40/56 | 458 | 1.472 (0.909–2.386) | 0.114 | 1.411 (0.381–2.395) | 0.202 |
| LSM14A | ||||||
| Low | 32/56 | 568 | 1 | 1 | ||
| High | 37/56 | 517 | 1.067 (0.660–1.726) | 0.792 | 0.814 (0.479–1.385) | 0.449 |
| LSM14B | ||||||
| Low | 35/56 | 517 | 1 | 1 | ||
| High | 34/56 | 592 | 0.994 (0.614–1.607) | 0.979 | 1.322 (0.772–2.263) | 0.309 |
Adjusted for histological grade, radiation therapy, radical resection, and targeted molecular therapy. MST, median survival time; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; TCGA, The Cancer Genome Atlas.
Combined effect survival analysis of LSM genes.
Patients were classified into high and low expression classes based on the expression of multiple LSM genes, and Table the expression frequency is shown in Table 2. Group C was shown to have poor OS compared with Groups A, B and D (adjusted P=0.001; adjusted HR =4.205, 95% CI: 1.778–9.948; Figure 6A). Using the Cox multivariable hazard regression model, regression coefficients of LSM1 and LSM4 were 0.719 and 0.346, respectively, and risk score = expression of (LSM1×0.719) + (expression of LSM4×0.346). The results were separated into different variables and classified as high-risk and low-risk groups by median cut off. The results showed that the high-risk level for patients with early-stage PDAC was positively correlated with a disappointing outcome (adjusted P=0.011; adjusted HR =2.038, 95% CI: 1.176–3.531; Table 3, Figure 6B and Figure 7A). Time-based ROC analysis showed that prognostic signatures successfully predicted the consequence of PDAC in early-stage patients (1-year area under the curve [AUC] =0.679; 2-year AUC =0.657; 5-year AUC =0.644; Figure 7B). The nomogram from this study also shows that LSM1 and LSM4 have a significant impact on the clinical outcomes of patients (Figure 7C). The proportions on nomogram rows represent the importance of each vector array, and the duration of row sections indicates how this element relates to the case.
Table 2
| Group | LSM1 | LSM4 | Events/total (n=112) | MST (days) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-value |
|---|---|---|---|---|---|---|---|---|
| A | Low | Low | 17/35 | 596 | 1 | 1 | ||
| B | Low | High | 13/21 | 470 | 2.390 (1.124–5.082) | 0.024 | 2.945 (1.270–6,826) | 0.012 |
| C | High | Low | 15/21 | 603 | 2.169 (1.051–4.474) | 0.036 | 4.205 (1.778–9.948) | 0.001 |
| D | High | High | 24/35 | 481 | 2.418 (1.263–4.629) | 0.008 | 3.327 (1.566–7.068) | 0.002 |
Adjusted for clinical variables, neoplasm histological grade, targeted molecular therapy, radiation therapy and residual resection. HR, hazard ratio; MST, median survival time; PLCD, phospholipase C delta; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; CI, confidence interval.
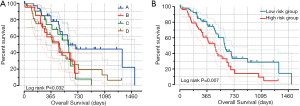
Table 3
| Risk scores | Events/total (n=112) | MST (days) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-value |
|---|---|---|---|---|---|---|
| Low | 32/56 | 607 | 1 | 1 | ||
| High | 37/56 | 473 | 1.932 (1.183–3.154) | 0.007 | 2.038 (1.176–3.531) | 0.011 |
Adjusted for histological grade, radiation therapy, radical resection, and targeted molecular therapy. MST, median survival time; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; TCGA, The Cancer Genome Atlas.
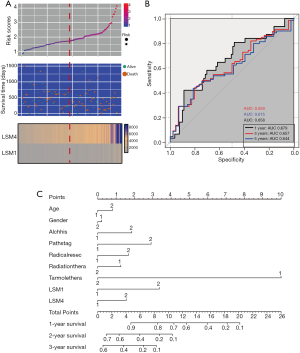
GSEA
Our study proved that LSM1 and LSM4 are significantly correlated with OS for patients with PDAC. Risk scores for LSM1 and LSM4 were combined and stratified according to the median. The GSEA results showed that the oncogenic mechanisms of LSM genes might include facilitating mRNA decapping, mRNA degradation, and mRNA splicing (Figure 8).

Discussion
The pancreatic tumor microenvironment comprises immune cells and cancer-associated fibroblasts (CAFs), which cause fibrosis of the interstitial connective tissue, and combined with the lack of angiogenesis in the pancreatic tumor, this results in a persistent and severely hypoxic tumor microenvironment (24,25). PDAC is a highly malignant tumor of the digestive system; the prognosis of patients with PDAC is extremely poor, and many patients are already at an advanced stage of cancer when diagnosed, thus early detection and early treatment are crucial for defeating PDAC (26). The standard diagnostic methods for PDAC included ultrasonography, computed tomography, positron emission tomography, magnetic resonance imaging, and endoscopic ultrasonography. However, because of the deep position of the pancreas, factors such as obesity and intestinal gas, and even the operator’s experience, can affect the accuracy of the diagnosis. Therefore, new biomarkers are urgently needed for detection of pancreatic cancer in the early stages (27). Existing markers with potentially important diagnostic value include carbohydrate antigen 19-9 (CA19-9), circulating tumor cells (CTCs), cell-free circulating tumor DNA (ctDNA), epigenetic biomarkers such as DNA methylation, histone modification, and chromatin structure, complement component 4 binding-protein a-chain (C4BPA), neutrophil gelatinase-associated lipocalin (NGAL) in urine, and others (28). However, CA19-9 is insufficient as an independent diagnostic tool; for instance, serum CA19-9 elevation can occur in patients with benign diseases such as chronic or acute pancreatitis, cholangitis, obstructive jaundice, liver cirrhosis, or other malignancies such as gastrointestinal cancers (29). CTCs are derived from primary tumors or metastatic sites and circulate in the bloodstream, and portal vein blood CTC counts but not peripheral blood CTC counts are correlated with OS (30). Neutrophil gelatinase-associated lipocalin requires numerous samples and multi-center clinical trials to identify and verify urinary diagnostic biomarkers of PDAC (28). PDAC is related to the oncogene K-ras, and downregulation of p53, p16/CDK2, and DPC4 (9).
LSM genes are significantly more highly expressed in pancreatic cancer than in surrounding normal tissues (11).LSM proteins are ubiquitous in eukaryotes, where they are present in both the nucleus and cytoplasm, and characterized by the Sm domain. LSMs are usually present as hexa- or heptamethyl RNA-binding complexes, forming two stable heptamer rings in LSM1-7 and LSM2-8 complexes, and their nuclear and cytoplasmic localization is tightly regulated. The two sturdy heptamer rings perform different functions; U6 snRNP contains an LSM2-8 heptamer, consisting of LSM2, LSM3, LSM4, LSM5, LSM6, LSM7, and LSM8, that resides in the nucleus and binds to the 3’ end of U6 snRNA and maintains its stability; meanwhile, the LSM1-7 heptamer is present in the cytoplasm and is involved in the degradation of mRNA (9,10,31-38). LSM proteins are involved in the occurrence and transformation of pancreatic cancer, and they are also related to changes in circadian rhythms in plants and mammals (9,39).
Using the TCGA database, we found that LSM genes are highly expressed in patients with pancreatic cancer. High expression levels of LSM1 and LSM4 were closely related to the prognosis of patients with PDAC. In pancreatic cancer patients with increased expression of LSM1 and LSM4, OS was significantly decreased. The multivariate Cox proportional risk regression model found that LSM1 and LSM4 may be early predictors of pancreatic cancer, and are essential target genes involved in the occurrence, proliferation, and metastasis of PDAC. Some previous studies found that the LSM1 gene is not only closely related to pancreatic cancer, but it could also provide a breakthrough in the treatment of this disease. However, only a few studies have explored the role of LSM1 in pancreatic tumor transformation and its possible mechanism, and not all members of the LSM gene family have been investigated. In the present work, we comprehensively explored the roles of all members of the LSM gene family in early-stage PDAC, and probed their potential molecular mechanisms.
Research has shown that LSM1 is essential for the transformation phenotype and anchorage-independent growth of pancreatic cancer cells; it can facilitate mRNA decapping and degradation, and thereby accelerate the cell cycle (40). In vitro experiments showed that LSM1 overexpression could induce the growth and cloning of pancreatic tumor cells. The possible molecular mechanism may involve LSM1 overexpression affecting the expression of various cancer-related pathways related to apoptosis and invasion, including transforming growth factor-β, tumor necrosis factor, and vascular endothelial growth factor, resulting in altered expression of Slug, MMP1, uPAR, and SerpinB5, all of which are known to be associated with invasion and metastasis in pancreatic cancer (11). Our GSEA results showed that LSM genes were related to RNA splicing, RNA synthesis, and RNA decomposition, and they may also indirectly cause carcinogenesis through other genes such as myc. Numerous studies have shown that upregulating MYC is a significant promoter of tumorigenesis, whether caused by other oncogenes (such as RAS mutations) or by MYC itself, and cell growth is correlated with MYC expression levels (41-43).
Progesterone can inhibit cancer cell cycle progression in PDAC and pancreatic neuroendocrine tumors (44-48). Additionally, patients with progesterone receptor-negative pancreatic solid pseudopapillary (SPNP) tumors have poor prognosis (49). Our current GSEA results confirmed that LSM3, LSM4, LSM7, and LSM8 might function by inhibiting progesterone. EZH2 expression is also upregulated in pancreatic cancer tissues, which can promote the proliferation and metastasis of pancreatic cancer cells (50-52), and our GSEA results indicate that LSM7 may play its role through EZH2.
The most important mechanism by which LSM proteins promote the development of cancer is through telomerase. A reverse transcriptase enzyme responsible for telomere elongation in cells is synthesized using a portion of the RNA subunit as a template, which can offset the loss of DNA sequences at the ends of chromosomes, since shortened telomeres have limited ability to replicate, and this enhances the proliferation capacity of cells in vitro (53-56). Studies have shown that many cancer cells display high telomerase activity in most eukaryotes, and the LSM2-8 complex can bind to the telomerase RNA subunit (TER1) of schizotypal yeast, and thereby protect mature TER1 and 3’ termini from external degradation, promoting the proliferation and malignant transformation of tumor cells (57).
Many patients are already at an advanced stage of cancer when diagnosed, and existing treatment methods such as chemotherapy, surgery, and radiation do not significantly improve the OS of patients with PDAC (58). The traditional standard treatment for PDAC is gemcitabine, and the most commonly used first-line treatment options are FOLFIRINOX (oxaliplatin + irinotecan +5-fluorouracil + leucovorin) or gemcitabine combined with nanoparticle albumin-bound (nab)-paclitaxel, all of which have limited efficacy in patients with advanced pancreatic cancer (59). Immunotherapy is useful for the treatment of leukemia and melanoma; it can activate host immune responses to identify and eliminate tumor cells. However, PDAC has a rich fibrotic stroma, and it lack lacks antigen-experienced T effector cells, hence immunotherapy is not effective for pancreatic cancer (60). The latest research shows that heat shock protein-90 can inhibit PSC/CAF in vitro and enhance the anti-PD-1 blocking effect in vivo (61). Additionally, in mouse experiments, combining anti-PD-1 and vaccine can have good anti-tumor activity; anti-CD137 agonist antibody combined with anti-PD-1 and vaccine significantly improved the survival time of the mouse PDAC model, highlighting a potential treatment option (62). Patients who received neoadjuvant and allogeneic PDAC vaccine tended to exhibit more prolonged median OS (63). Encouragingly, LSM1 antisense gene therapy can disrupt the tumor cell cycle, and this is very effective for pancreatic cancer. The growth of Panc02 tumors was significantly inhibited by injecting adenovirus-based LSM1 antisense RNA (Ad-Alsm1) into a subcutaneous tumor model (9,11). Similarly, the systematic application of Ad-ALSM1 in metastatic tumor models can substantially reduce tumor progression and prolong patient survival (11,40,64). LSM1 is also highly expressed in lung cancer, prostate cancer, breast cancer, and pancreatic endocrine tumors at both mRNA and protein levels. Antisense LSM1 transfection may reduce tumor cell proliferation via cyclin B1 and CDK1 proteins (40,65-69). Additionally, GSEA results in the present work indicate that LSM14B might help promote breast cancer progression through the ZNF217 gene.
The present study has some limitations. Firstly, the sample size was too small, and some patients are still being followed up. Thus, the probability of false-negative results is high, and more specimens are needed for further verification. Secondly, the clinical information is not comprehensive enough, and neither are the results. Thirdly, performed analysis only at the transcriptional level. Nonetheless, we discovered new ties between LSM genes and predictions of early PDAC, but further investigation is needed to confirm these results and the fundamental molecular mechanisms.
Despite these limitations, an association between LSM genes and early PDAC patient prognosis was demonstrated for the first time. Additionally, GSEA was used to assess the possible molecular mechanisms by which LSM genes may influence the prognostics of patients with early PDC. Moreover, LSM genes are likely to become new targets for PDAC therapy if future studies confirm these observations.
Conclusions
In conclusion, high expression levels of LSM1 and LSM4 is related to poor prognosis in early-stage PDAC patients, and these genes may serve as potential prognostic biomarkers and therapeutic targets in early-stage PDAC.
Acknowledgments
The authors thank TCGA Shared database (https://cancergenome.nih.gov/) in patients with pancreatic cancer RNA sequencing data, also thank the university of California, Santa Cruz, Xena (UCSC Xena http://xena.ucsc.edu/) sharing these patients with the corresponding clinical data and survival. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: The present study was partly supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3056
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3056). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All data used in this study were downloaded from a public database, and no humans or animals were involved in this research. Hence, no approval from an ethics committee was required. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mishra NK, Southekal S, Guda C. Survival Analysis of Multi-Omics Data Identifies Potential Prognostic Markers of Pancreatic Ductal Adenocarcinoma. Front Genet 2019;10:624. [Crossref] [PubMed]
- Young K, Hughes DJ. Immunotherapy and pancreatic cancer: unique challenges and potential opportunities 2018;10:1758835918816281.
- Swords DS, Firpo MA, Scaife CL, Mulvihill SJ. Biomarkers in pancreatic adenocarcinoma: current perspectives. Onco Targets Ther 2016;9:7459-67. [Crossref] [PubMed]
- Krška Z, Šváb J, Hoskovec D, et al. Pancreatic Cancer Diagnostics and Treatment--Current State. Prague Med Rep 2015;116:253-67. [Crossref] [PubMed]
- Rouanet M, Lebrin M, Gross F, et al. Gene Therapy for Pancreatic Cancer: Specificity, Issues and Hopes. Int J Mol Sci 2017;18:1231. [Crossref] [PubMed]
- Xue R, Chen L, Zhang C, et al. Genomic and Transcriptomic Profiling of Combined Hepatocellular and Intrahepatic Cholangiocarcinoma Reveals Distinct Molecular Subtypes. Cancer Cell 2019;35:932-47.e8. [Crossref] [PubMed]
- Sun T, Kong X, Du Y, Li Z. Aberrant MicroRNAs in Pancreatic Cancer: Researches and Clinical Implications. Gastroenterol Res Pract 2014;2014:386561. [Crossref] [PubMed]
- Xue R, Hua L, Xu W, et al. Derivation and Validation of the Potential Core Genes in Pancreatic Cancer for Tumor-Stroma Crosstalk. Biomed Res Int 2018;2018:4283673. [Crossref] [PubMed]
- Yan Y, Rubinchik S, Wood AL, et al. Bystander effect contributes to the antitumor efficacy of CaSm antisense gene therapy in a preclinical model of advanced pancreatic cancer. Mol Ther 2006;13:357-65. [Crossref] [PubMed]
- Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, et al. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. The Plant cell 2012;24:4930-47. [Crossref] [PubMed]
- Little EC, Camp ER, Wang C, et al. The CaSm (LSm1) oncogene promotes transformation, chemoresistance and metastasis of pancreatic cancer cells. Oncogenesis 2016;5:e182. [Crossref] [PubMed]
- Yan Y, Rubinchik S, Watson PM, et al. Establishing a murine pancreatic cancer CaSm model: up-regulation of CaSm is required for the transformed phenotype of murine pancreatic adenocarcinoma. Mol Ther 2005;11:363-72. [Crossref] [PubMed]
- Schweinfest CW, Graber MW, Chapman JM, et al. CaSm: an Sm-like protein that contributes to the transformed state in cancer cells. Cancer Res 1997;57:2961-5. [PubMed]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35:W169-75. [Crossref] [PubMed]
- Montojo J, Zuberi K, Rodriguez H, et al. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res 2014;3:153. [Crossref] [PubMed]
- Liao X, Huang K, Huang R, et al. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther 2017;10:4493-506. [Crossref] [PubMed]
- Liao X, Zhu G, Huang R, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res 2018;10:787-803. [Crossref] [PubMed]
- Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics 2005;61:92-105. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- François O, Martins H, Caye K, et al. Controlling false discoveries in genome scans for selection. Mol Ecol 2016;25:454-69. [Crossref] [PubMed]
- Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol 2014;67:850-7. [Crossref] [PubMed]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003;19:368-75. [Crossref] [PubMed]
- Sunami Y, Häußler J, Kleeff J. Cellular Heterogeneity of Pancreatic Stellate Cells, Mesenchymal Stem Cells, and Cancer-Associated Fibroblasts in Pancreatic Cancer. Cancers (Basel) 2020;12:3770. [Crossref] [PubMed]
- Tao J, Yang G, Zhou W, et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol 2021;14:14. [Crossref] [PubMed]
- Sivapalan L, Kocher HM, Ross-Adams H, et al. Molecular profiling of ctDNA in pancreatic cancer: Opportunities and challenges for clinical application. Pancreatology 2021;21:363-78. [Crossref] [PubMed]
- Li S, Wang L, Zhao Q, Wang Z, et al. Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer. Front Genet 2020;11:596078. [Crossref] [PubMed]
- Zhang WH, Wang WQ, Han X, et al. Advances on diagnostic biomarkers of pancreatic ductal adenocarcinoma: A systems biology perspective. Comput Struct Biotechnol J 2020;18:3606-14. [Crossref] [PubMed]
- Lee T, Teng TZJ, Shelat VG. Carbohydrate antigen 19-9 - tumor marker: Past, present, and future. World J Gastrointest Surg 2020;12:468-90. [Crossref] [PubMed]
- Bali P, Lozano-Pope I, Pachow C, et al. Early detection of tumor cells in bone marrow and peripheral blood in a fast-progressing gastric cancer model. Int J Oncol 2021;58:388-96. [Crossref] [PubMed]
- Chowdhury A, Raju KK, Kalurupalle S, et al. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1-7-Pat1 complex. RNA 2012;18:936-44. [Crossref] [PubMed]
- Kufel J, Bousquet-Antonelli C, Beggs JD, et al. Nuclear pre-mRNA decapping and 5' degradation in yeast require the Lsm2-8p complex. Mol Cell Biol 2004;24:9646-57. [Crossref] [PubMed]
- Spiller MP, Reijns MA, Beggs JD. Requirements for nuclear localization of the Lsm2-8p complex and competition between nuclear and cytoplasmic Lsm complexes. J Cell Sci 2007;120:4310-20. [Crossref] [PubMed]
- Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Molecular cell 2001;8:1075-83. [Crossref] [PubMed]
- Lyons SM, Ricciardi AS, Guo AY, et al. The C-terminal extension of Lsm4 interacts directly with the 3' end of the histone mRNP and is required for efficient histone mRNA degradation. Rna 2014;20:88-102. [Crossref] [PubMed]
- Reimer KA, Stark MR, Aguilar LC, et al. The sole LSm complex in Cyanidioschyzon merolae associates with pre-mRNA splicing and mRNA degradation factors. RNA 2017;23:952-67. [Crossref] [PubMed]
- He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3' termini from partial degradation. Genetics 2001;158:1445-55. [PubMed]
- Zaric B, Chami M, Rémigy H, et al. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem 2005;280:16066-75. [Crossref] [PubMed]
- Perez-Santangelo S, Mancini E, Francey LJ, et al. Role for LSM genes in the regulation of circadian rhythms. Proc Natl Acad Sci U S A 2014;111:15166-71. [Crossref] [PubMed]
- Fraser MM, Watson PM, Fraig MM, et al. CaSm-mediated cellular transformation is associated with altered gene expression and messenger RNA stability. Cancer Res 2005;65:6228-36. [Crossref] [PubMed]
- Dang CV. MYC on the path to cancer. Cell 2012;149:22-35. [Crossref] [PubMed]
- Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J 2017;36:3409-20. [Crossref] [PubMed]
- Rickman DS, Schulte JH, Eilers M. The Expanding World of N-MYC-Driven Tumors. Cancer Discov 2018;8:150-63. [Crossref] [PubMed]
- Yazdani S, Kasajima A, Onodera Y, et al. Progesterone arrested cell cycle progression through progesterone receptor isoform A in pancreatic neuroendocrine neoplasm. J Steroid Biochem Mol Biol 2018;178:243-53. [Crossref] [PubMed]
- Goncharov AI, Maslakova AA, Polikarpova AV, et al. Progesterone inhibits proliferation and modulates expression of proliferation-Related genes in classical progesterone receptor-negative human BxPC3 pancreatic adenocarcinoma cells. J Steroid Biochem Mol Biol 2017;165:293-304. [Crossref] [PubMed]
- Estrella JS, Broaddus RR, Mathews A, et al. Progesterone receptor and PTEN expression predict survival in patients with low- and intermediate-grade pancreatic neuroendocrine tumors. Arch Pathol Lab Med 2014;138:1027-36. [Crossref] [PubMed]
- Viale G, Doglioni C, Gambacorta M, et al. Progesterone receptor immunoreactivity in pancreatic endocrine tumors. An immunocytochemical study of 156 neuroendocrine tumors of the pancreas, gastrointestinal and respiratory tracts, and skin. Cancer 1992;70:2268-77. [Crossref] [PubMed]
- Yazdani S, Kasajima A, Ogata H, et al. Progesterone Receptor Isoforms A and B in Pancreatic Neuroendocrine Tumor. Neuroendocrinology 2015;101:309-20. [Crossref] [PubMed]
- Wang F, Meng Z, Li S, et al. Prognostic value of progesterone receptor in solid pseudopapillary neoplasm of the pancreas: evaluation of a pooled case series. BMC Gastroenterol 2018;18:187. [Crossref] [PubMed]
- Bao B, Ali S, Banerjee S, et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res 2012;72:335-45. [Crossref] [PubMed]
- Mallen-St Clair J, Soydaner-Azeloglu R, Lee KE, et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev 2012;26:439-44. [Crossref] [PubMed]
- Chen Y, Xie D, Yin Li W, et al. RNAi targeting EZH2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo. Cancer Lett 2010;297:109-16. [Crossref] [PubMed]
- Bernardes de Jesus B, Blasco MA. Telomerase at the intersection of cancer and aging. Trends Genet 2013;29:513-20. [Crossref] [PubMed]
- Parks JW, Stone MD. Single-Molecule Studies of Telomeres and Telomerase. Annu Rev Biophys 2017;46:357-77. [Crossref] [PubMed]
- Chen L, Roake CM, Freund A, et al. An Activity Switch in Human Telomerase Based on RNA Conformation and Shaped by TCAB1. Cell 2018;174:218-30.e13. [Crossref] [PubMed]
- Arndt GM, MacKenzie KL. New prospects for targeting telomerase beyond the telomere. Nat Rev Cancer 2016;16:508-24. [Crossref] [PubMed]
- Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 2012;484:260-4. [Crossref] [PubMed]
- Ansari D, Gustafsson A, Andersson R. Update on the management of pancreatic cancer: surgery is not enough. World J Gastroenterol 2015;21:3157-65. [Crossref] [PubMed]
- Robinson CM, Talty A, Logue SE, et al. An Emerging Role for the Unfolded Protein Response in Pancreatic Cancer. Cancers (Basel) 2021;13:261. [Crossref] [PubMed]
- Tran TH, Mattheolabakis G, Aldawsari H, et al. Exosomes as nanocarriers for immunotherapy of cancer and inflammatory diseases. Clin Immunol 2015;160:46-58. [Crossref] [PubMed]
- Zhang Y, Ware MB, Zaidi MY, et al. Heat Shock Protein-90 Inhibition Alters Activation of Pancreatic Stellate Cells and Enhances the Efficacy of PD-1 Blockade in Pancreatic Cancer. Mol Cancer Ther 2021;20:150-60. [Crossref] [PubMed]
- Muth ST, Saung MT, Blair AB, et al. CD137 agonist-based combination immunotherapy enhances activated, effector memory T cells and prolongs survival in pancreatic adenocarcinoma. Cancer Lett 2021;499:99-108. [Crossref] [PubMed]
- Zheng L, Ding D, Edil BH, et al. Vaccine-Induced Intratumoral Lymphoid Aggregates Correlate with Survival Following Treatment with a Neoadjuvant and Adjuvant Vaccine in Patients with Resectable Pancreatic Adenocarcinoma. Clin Cancer Res 2021;27:1278-86. [Crossref] [PubMed]
- Kelley JR, Brown JM, Frasier MM, et al. The cancer-associated Sm-like oncogene: a novel target for the gene therapy of pancreatic cancer. Surgery 2000;128:353-60. [Crossref] [PubMed]
- Watson PM, Miller SW, Fraig M, et al. CaSm (LSm-1) overexpression in lung cancer and mesothelioma is required for transformed phenotypes. Am J Respir Cell Mol Biol 2008;38:671-8. [Crossref] [PubMed]
- Gumbs AA, Bassi C, Moore PS, et al. Overexpression of the Sm-like proto-oncogene in primary and metastatic pancreatic endocrine tumors. Jop 2002;3:109-15. [PubMed]
- Streicher KL, Yang ZQ, Draghici S, et al. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene 2007;26:2104-14. [Crossref] [PubMed]
- Yang ZQ, Liu G, Bollig-Fischer A, et al. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res 2010;70:8487-97. [Crossref] [PubMed]
- Takahashi S, Suzuki S, Inaguma S, et al. Down-regulation of Lsm1 is involved in human prostate cancer progression. Br J Cancer 2002;86:940-6. [Crossref] [PubMed]



