
Bioinformatics analysis reveals that ANXA1 and SPINK5 are novel tumor suppressor genes in patients with oral squamous cell carcinoma
Introduction
Malignant tumors are one of the main threats to national public health and the second most common cause of death. It is expected that there will be more than 20 million new cases of malignant tumors in 2025 (1). Among all malignant tumors, squamous cell carcinoma of the head and neck (HNSCC) is the sixth most common malignant tumor worldwide and includes oral squamous cell carcinoma (OSCC) (2,3). In 2012, the mortality rate of 145,000 patients with OSCC was approximately 1.8% (4). Interestingly, the rate of male patients with OSCC is particularly high, with the incidence ranging from 1 per 100,000 to 10 per 100,000 (5). It is accepted that adopting unhealthy lifestyles, such as smoking, poor diet, and lack of exercise increases the risk of OSCC (6,7). According to statistics, the incidence of OSCC in different countries and regions is also different. For example, in Central and South Asia, OSCC is one of the three most common malignant tumors. In India, the age-standardized incidence of OSCC is 12.6 per 100,000 population. In addition, the incidence of OSCC in several countries and regions, including Denmark, France, Germany, Scotland, Central, and Eastern Europe, has increased sharply (4).
OSCC can occur in different locations, being present in squamous epithelium and scattered in smaller salivary glands and lymphatic drainage pathways. Consequently, OSCC cells can easily invade tissues and enter the cervical lymph nodes. Induction of epithelial-mesenchymal transition (EMT) in squamous cell carcinoma promotes tumor development by enhancing the characteristics of cell invasion and metastasis (8). Oncogenes and tumor suppressor genes are the two main gene groups that regulate tumorigenesis, promoting tumorigenesis when they are expressed at high and low levels, respectively. Due to the lack of control mechanisms for cell growth and differentiation, the development of OSCC is related to changes in the expression of related genes. The development of tumors is attributed to a series of genetic events, including the abnormal activity of oncogenes, related invasion genes, and inactivation of tumor suppressor genes.
Tobacco consumption and excessive drinking account for about 90% of the causes of oral malignancies and when combined with alcohol or betel nut consumption (9), the risk of OSCC increases (10). Smokers have long-term exposure to many toxic ingredients, including toxic metals (11). Since neither oral epithelium nor lung epithelium constitutes a substantial barrier to these toxic substances (12), small particles and soluble toxic substances can be absorbed into the epithelium and interstitial tissues (13). The accumulation of acetaldehyde in human tissues depends on the balance between the rate of formation of alcohol dehydrogenase and acetaldehyde, and the rate of removal of aldehyde dehydrogenase. There is a significant association between low-activity aldehyde dehydrogenase and the risk of OSCC in East Asian heavy drinkers (13).
Currently, the main treatment strategy for OSCC is still surgery. However, since most patients have advanced tumors when diagnosed, radiotherapy and chemotherapy are important adjuvant treatments for OSCC. For patients with locally advanced OSCC who are at high risk of recurrence or metastasis, multiple treatment strategies may improve their prognosis. However, treatment strategies are still limited, with further studies being needed to explore new potential biomarkers for prognosis and novel treatment targets for precision medicine.
Based on high-throughput gene expression databases, the current study explores the potential molecular mechanisms of OSCC, to identify differentially-expressed genes (DEGs) in OSCC and their regulatory relationships in order to elucidate the molecular basis of OSCC and provide a theoretical basis and individualized precise therapeutic targets for the treatment of OSCC patients. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3382).
Methods
Microarray datasets
Datasets were searched in the GEO database, using OSCC as the keyword. To compare the different gene expression patterns between cancer and normal tissues, the studies using OSCC patients and normal individuals were included and research focusing on OSCC cells was excluded. Three GEO datasets, GSE146483, GSE138206 and GSE148944 were downloaded.
GSE146483 included 8 OSCC tissues and 3 normal oral mucosal epithelial keratinocytes. GSE138206 was obtained from the microarray data of 6 OSCC tissues, tissues adjacent to cancer, and contralateral normal tissues. GSE148944 was collected from a retrospective, cohort study, including up to 40 OSCC cases and 6 non-cancer controls (14). The clinicopathological variables of oral cancer, to identify and validate potentially implementable genes, were analyzed from GSE84846 (15), which provided the age of diagnosis, gender, primary tumor stage and lymph node stage of OSCC patients.
Identification of DEGs
For analyzing differential expression in the GEO datasets, the limma package of R software, GEO2R, was used to analyze the original datasets, with a cutoff value |log FC| >1 and P-value <0.01. Volcano maps were constructed by SangerBox software. The DEGs common to all three datasets were identified, by using Venn package (http://bioinformatics.psb.ugent.be/webtools/Venn/), to find biomarkers or treatment targets of OSCC with the most potential.
Expression pattern of DEGs
An online cancer microarray database-mining platform, the Oncomine database (https://www.oncomine.org/), was used to investigate the differences in transcriptional levels of the DEGs in malignant tumors and normal tissues. The protein level of DEGs was analyzed using the Human Protein Atlas (https://www.proteinatlas.org/) to obtain the expression levels of DEGs in normal tissues and OSCC tumor tissues.
Relationship between DEG expression and the clinicoparameters of OSCC patients
As DEGs are potential biomarkers or therapeutic targets of OSCC, their expression may affect the progress of OSCC development. To explore the relationship between the expression of DEGs and the clinicoparameters of OSCC patients, the GSE84846 dataset was downloaded, including the value of DEG expression and the clinical information. The median value was set as the cutoff for low/high expression of DEGs. Chi-square analysis was used to analyze their relationship and a P<0.05 was considered as statistically significant.
Mutation patterns of DEGs
As mutation occurs frequently, playing an important role in the oncogenesis and development of malignant tumors, Oral Squamous Cell Carcinoma (MD Anderson, Cancer Discov 2013) in the cBioPortal dataset (www.cbioportal.org/), was used to analyze the mutation patterns of DEGs in OSCC.
Survival analysis according to DEGs in OSCC patients
To explore the prognostic value of the OSCC DEGs, the Human Protein Atlas (www.proteinatlas.org/) was used to analyze the impact of DEGs on survival of OSCC patients, while the cBioPortal database (http://www.cbioportal.org/) for multi-dimensional analysis of data from The Cancer Genome Atlas (TCGA) was used to analyze the effects of mutations in DEGs on the survival of patients with OSCC.
Statistical analysis and ethic statement
Levels of statistical significance were analyzed with data by using the Chi-square test for categorical variables. All statistical difference were considered significant at the level of P<0.05, conducted with SPSS 16.0 for windows. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Gene expression differences between normal and OSCC tissues
Based on the search results, three microarray datasets of gene expression, GSE146483, GSE138206, and GSE148944 were downloaded from the GEO database of normal and OSCC tissues. A volcano map indicated the potential DEGs in OSCC tissues (Figure 1A,B,C). The intersection between GSE146483, GSE138206, and GSE148944 was comprised of 7 DEGs, specifically CXCR4, ANXA1, MSR1, IL1RN, SPINK5, APOE, and IFI35 (Figure 1D).

Expression of the DEGs in malignant tumors
The Oncomine dataset provides extensive expression information of several genes in different malignant tumors. Expression patterns of the 7 DEGs in different cancers were diverse. For HNSCC, including OSCC, the expression of CXCR4, APOE, IFI35, and MSR1 was increased in cancer tissues compared with normal tissues, whereas down-regulated expression of SPINK5, IL1RN and ANXA1 was found in cancer tissues compared to normal tissues (Figure 2).
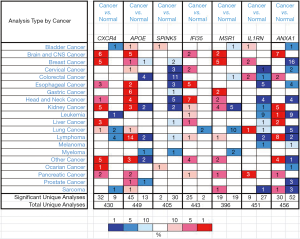
The increased DEGs in OSCC tissues
To investigate the potential function of the DEGs in OSCC, further studies were explored and analyzed for their expression in different tissues. However, as research on OSCC is limited, only the expression of CXCR4, IFI35, and MSR1 in tumor tissues was higher than that in normal tissues, showing 3.447, 3.104, and 3.432-fold increases, respectively (Figure 3).
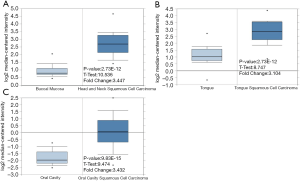
The protein level of DEGs in OSCC and normal tissues
To further analyze the protein levels of the DEGs in tumor tissues, the Human Protein Atlas was used to analyze the expression and location of DEGs in normal tissues and malignant tumors. ANXA1, IL1RN, SPINK5, and APOE were down-regulated, and IFI35 was up-regulated in cancer tissues (Figure 4), supporting our DEG analysis. Protein expression of MSR1 was negative in normal and cancer tissues both (Figure 4C,4D). No report on the protein level of CXCR4 in OSCC or normal tissues was found in the Human Protein Atlas.
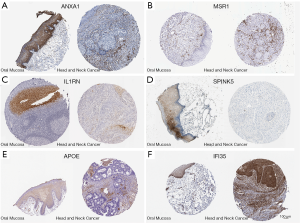
Expression of the DEGs is associated with TNM stage and node status in OSCC patients
To verify the potential function of DEGs in OSCC, information for the 99 OSCC patients in the GSE84846 dataset was downloaded and analyzed. The age range was from 28 to 89, and the sex ratio was approximately 1:1. In Table 1, low expression of ANXA1 was positively associated with the early stage of TNM (P=0.012), and the expression of SPINK5 was associated with the TNM stage and node status, with P-values 0.001 and 0.011, respectively.
Table 1
| Gene | TNM | N | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I–III | % | IV | % | P value | 0 | % | 1–2 | % | P-value | ||
| CXCR4 | |||||||||||
| Low | 25 | 51 | 24 | 49 | 0.369 | 21 | 44 | 27 | 56 | 0.264 | |
| High | 30 | 60 | 20 | 40 | 27 | 55 | 22 | 45 | |||
| ANXA1 | |||||||||||
| Low | 21 | 43 | 28 | 57 | 0.012 | 20 | 41 | 29 | 59 | 0.084 | |
| High | 34 | 68 | 16 | 32 | 28 | 58 | 20 | 42 | |||
| MSR1 | |||||||||||
| Low | 30 | 61 | 19 | 39 | 0.261 | 25 | 52 | 23 | 48 | 0.612 | |
| High | 25 | 50 | 25 | 50 | 23 | 47 | 26 | 53 | |||
| ILIRN | |||||||||||
| Low | 24 | 49 | 25 | 51 | 0.192 | 22 | 46 | 26 | 54 | 0.477 | |
| High | 31 | 62 | 19 | 38 | 26 | 53 | 23 | 47 | |||
| SPINK5 | |||||||||||
| Low | 19 | 39 | 30 | 61 | 0.001 | 18 | 37 | 31 | 63 | 0.011 | |
| High | 36 | 72 | 14 | 28 | 30 | 63 | 18 | 37 | |||
| IFI35 | |||||||||||
| Low | 29 | 59 | 20 | 41 | 0.472 | 23 | 49 | 24 | 51 | 0.917 | |
| High | 26 | 52 | 24 | 48 | 25 | 50 | 25 | 50 | |||
DEGs, differentially-expressed genes; OSCC, oral squamous cell carcinoma.
No mutations in DEGs were found in OSCC patients
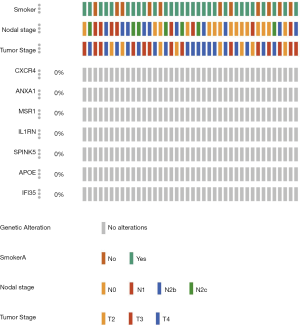
Although mutation occurs frequently in different malignant tumors, no mutations in the 7 common DEGs were found in OSCC patients, regardless of patient smoking, or the nodal or tumor stage (Figure 5), suggesting that the DEG expression pattern rather than mutation of the DEGs plays the important role in oncogenesis and development of OSCC.
Survival value of DEGs in OSCC (Figure 6)
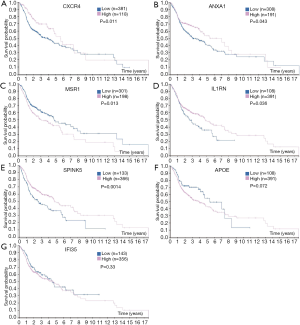
Besides the expression and location of DEGs in tissues, the Human Protein Atlas also provides the prognostic value of genes in different cancers. High expression of MSR1 in OSCC patients was prognostic of poor overall survival (OS) (P=0.0014; Figure 6C), whereas low expression of CXCR4, ANXA1, IL1RN, and SPINK5 predicted poor OS in OSCC patients (P<0.01) (Figure 6A,B,D,E). Although the expression of other DEGs did not show a significant relationship with the survival of OSCC patients (P>0.05), OSCC patients with low expression of APOE (Figure 6F) tended to have longer lifespans.
Discussion
HNSCC are common in elderly people with a history of exposure to tobacco and alcohol (16). Tobacco smoke is a mixture of chemical substances, of which there are at least 60 substances that may cause malignant tumors or cause somatic mutations by inducing DNA damage. These substances may also cause driver mutations in malignant tumor-related genes (17). Research on the genome of malignant tumors associated with smoking can deepen the understanding of how this risk factor can lead to malignant tumors (18). However, in the current study, no mutations were found in the common DEGs we identified for OSCC patients, suggesting that the regulation of DEG expression is important for the oncogenesis and development of OSCC.
Currently, the survival rate for oral cancer is very low. Despite progress in treatment intervention, the survival rate has not improved significantly in recent decades (19).Variables, such as age, co-morbidity, immune and nutritional status, tumor size and location, lymph node status, oncogene expression, proliferation markers, or DNA content, have become independent prognostic markers for evaluating oral cancer (20). The advanced stages of squamous cell carcinoma are often associated with high mortality (21). Early detection of oral cancer is the most effective means to reduce the mortality and morbidity of the disease, but progress in this field has been slow (22). Increasing evidence indicates that oral examination of high-risk groups may be a cost-effective screening strategy (23). The determinants of diagnosis delay (the time elapsed from the first sign or symptom to the final diagnosis) have been reported (24). Up to now, it is not practical to screen for oral cancer during each routine check-up, and selective screening is a more realistic and effective solution. Unfortunately, most malignant tumors, when diagnosed, are advanced (third or fourth stage), and the 5-year survival rate is between 10% and 40% (25). The diagnosis rate of ulcerated OSCC in the third to fourth stages can be as high as 60%, but this correlation does not reach statistical significance (26). In addition, although ulcerative lesions indicate poor survival, the predictive value of clinical survival is still controversial.
To explore further potential biomarkers for OSCC, high-throughput gene expression datasets were recruited and 7 potential DEGs, common to all datasets, were identified in the current study. The chemokine receptor CXCR4 is a specific receptor for chemokine stromal cell-derived factor-1 (CXCL12) (27). Blocking CXCR4 significantly reduces chemotaxis of M2 macrophages, and polarized M2 macrophages promote CSC-like transition in OSCC cell lines (27). The CXCL12-CXCR4/CXCR7 axis contributes to cell motility in OSCC (28). Interestingly, hypoxia, closely related to malignant tumors, enhances CXCR4 expression in OSCC through activating HIF-1 (29). Meanwhile, targeted silencing CXCR4 expression significantly inhibits the EMT process of OSCC (30). As the predicting role of low CXCR4 mRNA level is found in the current study, further exploration is needed to investigate the underlying mechanism of CXCR4 in OSCC.
Annexin A1 (ANXA1) protein has been suggested to be an active player in the tumorigenesis of many organs, and a tumor suppressor role for Annexin A1in OSCC is shown in the current study as well as several prior investigations. Faria et al. first reported down-regulated ANXA expression at the mRNA level in OSCC and correlates with age, sex, and the anatomical site of the tumor lesion (31). Interestingly, loss of plasma membrane ANXA1 expression is significantly correlated with a poorly differentiated status of OSCC cells, which is frequent during oral carcinogenesis (32).On the other hand, Lin et al. reported the nuclear localization of ANXA1 as a prognostic factor of OSCC and occurs frequently (33). Consistently, current research has predicted ANXA1 to be a novel tumor suppressor gene in the oncogenesis of OSCC, and may be related EMT (34). Furthermore, TGFβ1/EGF-induced EMT in OSCC cell lines is attenuated by ANXA1 overexpression and TGFβ1/EGF-induced EMT can be reversed by ANXA1 in OSCC (35), suggesting ANXA1 to be a potential marker as well as novel therapeutic target for OSCC.
MSR1 repeats are a 36-38 bp minisatellite element that has recently been implicated in the regulation of gene expression, through copy number variation (CNV), and act as molecular switches to modulate gene expression (36). Up to now, no report has shown a relationship between OSCC and MSR1, which is usually associated with hereditary prostate cancer (37). Interestingly, low expression of MSR1 without mutation predicts long overall survival, serving as a prognostic biomarker for OSCC patients.
Interleukin-1 receptor antagonist (IL1RN), a potent anti-inflammatory molecule, modulates the biological activity of the pro-inflammatory cytokine, interleukin-1. After investigating the expression of IL1RN in OSCC, Shiiba et al. reported that IL1RN may exhibit opposing characteristics in oral malignancies, depending on the stage of cancer development, suppressing early carcinogenic events, yet promoting tumor development in some lesion sites (38). Shan et al. reported that salivary proteomics is promising for the discovery of OSCC biomarkers (39). In the microarray dataset, expression of IL1RN was also found to be downregulated in OSCC tissues compared with normal tissue and predicts poor overall survival of patients.
Serine protease inhibitor Kazal-type 5 (SPINK5) is an inhibitor of kallikrein (KLK) 5 and KLK7, which have been recently validated in a lymph node metastasis-predicting gene expression profile in OSCC (40). The SPINK5 gene provides instructions for making a protein called LEKT1, a serine peptidase inhibitor, controlling the activity of enzymes to break down other proteins (40). Zhao et al. demonstrated that SPINK5 is significantly associated with the survival of OSCC patients, and its differential expression in OSCC patients has been further confirmed by reverse transcription-quantitative polymerase chain reaction in OSCC and normal oral cell lines (41). The SPINK5 gene caused by epigenetic changes via DNA methylation may be associated with the development and progression of OSCC (41). Consistently, the findings in this study show the prognostic value of SPINK5 in OSCC patients, with low expression patterns being closely associated with TNM stage and node status.
Apolipoprotein E (APOE) combines with fats (lipids) in the body to form lipoproteins, responsible for packaging cholesterol and other fats and carrying them through the bloodstream (42). Maintaining normal levels of cholesterol is essential for the prevention of disorders that affect the heart and blood vessels. Importantly, abnormal APOE expression is also related to the development of malignant tumors, including melanoma (43), and gastric cancer (44). However, research focused on APOE in OSCC is limited. Jayakar et al. investigated the function of APOE in OSCC and found that APOE can promote the invasion of OSCC cells through regulating the levels of phospho-extracellular signal-regulated kinase 1/2, phospho-c-Jun N-terminal kinase, phospho-c-Jun and activator protein 1 (AP-1) (45). Further investigation needs to explore the underlying molecular mechanism.
Interferon-induced protein 35 (IFI35), an important element of the cellular response to viral infection, has been suggested to regulate the innate immune response (46). Up to now, the function of IFI35 on the development of malignant tumors is still unclear. However, Shirai et al. reported IFI35 negatively regulates the TLR3-IFN-β-P-STAT1-RIG-I-CXCL10/CCL5 axis in U373MG cells (47), at least partially uncovering the molecular mechanism of IFI35 in tumors and forging a relationship between IFI35 and cytokines, which is significantly associated with the development of malignant tumors. In conclusion, our findings uncover 7 potential biomarkers common to OSCC patients, in particular identifying ANXA1 and SPINK5 as novel tumor suppressor genes in OSCC, to provide new treatment strategies for precision medicine in individual patients.
Acknowledgments
We are thankful to Prof. Shanley Lin for his critical and careful editing and proofreading of the manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3382
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3382) and reports grants from National Natural Science Foundation of China, during the conduct of the study. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ligiero Braga T, Santos-Oliveira R. PPoma Review: Epidemiology, Aetiopathogenesis, Prognosis and Treatment. Diseases 2018;6:8. [Crossref] [PubMed]
- Kim SY, Nam SY, Choi SH, et al. Prognostic value of lymph node density in node-positive patients with oral squamous cell carcinoma. Ann Surg Oncol 2011;18:2310-7. [Crossref] [PubMed]
- Ong W, Zhao R, Lui B, et al. Prognostic significance of lymph node density in squamous cell carcinoma of the tongue. Head Neck 2016;38:E859-66. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Laniosz V, Torgerson RR, Ramos-Rodriguez AJ, et al. Incidence of squamous cell carcinoma in oral lichen planus: a 25-year population-based study. Int J Dermatol 2019;58:296-301. [Crossref] [PubMed]
- World Health Organization. World Health Assembly adopts historic tobacco control pact. Indian J Med Sci 2003;57:377-8. [PubMed]
- World Health Assembly adopts landmark strategies on diet and physical activity, and reproductive health. Bull World Health Organ 2004;82:478. [PubMed]
- Wang J, Jiang C, Li N, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis 2020;11:682. [Crossref] [PubMed]
- Lee CH, Ko AM, Warnakulasuriya S, et al. Intercountry prevalences and practices of betel-quid use in south, southeast and eastern Asia regions and associated oral preneoplastic disorders: an international collaborative study by Asian betel-quid consortium of south and east Asia. Int J Cancer 2011;129:1741-51. [Crossref] [PubMed]
- Reibel J. Tobacco and oral diseases. Update on the evidence, with recommendations. Med Princ Pract 2003;12:22-32. [Crossref] [PubMed]
- Pappas RS, Stanfill SB, Watson CH, et al. Analysis of toxic metals in commercial moist snuff and Alaskan iqmik. J Anal Toxicol 2008;32:281-91. [Crossref] [PubMed]
- Mauro M, Crosera M, Bianco C, et al. In vitro permeability of silver nanoparticles through porcine oromucosal membrane. Colloids Surf B Biointerfaces 2015;132:10-6. [Crossref] [PubMed]
- Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health 2006;29:245-54. [PubMed]
- Meehan K, Leslie C, Lucas M, et al. Characterization of the immune profile of oral tongue squamous cell carcinomas with advancing disease. Cancer Med 2020;9:4791-807. [Crossref] [PubMed]
- Mes SW, Te Beest D, Poli T, et al. Prognostic modeling of oral cancer by gene profiles and clinicopathological co-variables. Oncotarget 2017;8:59312-23. [Crossref] [PubMed]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. [Crossref] [PubMed]
- Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 2003;3:733-44. [Crossref] [PubMed]
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60. [Crossref] [PubMed]
- Taghavi N, Yazdi I. Prognostic factors of survival rate in oral squamous cell carcinoma: clinical, histologic, genetic and molecular concepts. Arch Iran Med 2015;18:314-9. [PubMed]
- Llewellyn CD, Johnson NW, Warnakulasuriya KA. Risk factors for oral cancer in newly diagnosed patients aged 45 years and younger: a case-control study in Southern England. J Oral Pathol Med 2004;33:525-32. [Crossref] [PubMed]
- Johnson NW, Warnakulasuriy S, Tavassoli M. Hereditary and environmental risk factors; clinical and laboratory risk matters for head and neck, especially oral, cancer and precancer. Eur J Cancer Prev 1996;5:5-17. [PubMed]
- Baykul T, Yilmaz HH, Aydin U, et al. Early diagnosis of oral cancer. J Int Med Res 2010;38:737-49. [Crossref] [PubMed]
- McGurk M, Scott SE. The reality of identifying early oral cancer in the general dental practice. Br Dent J 2010;208:347-51. [Crossref] [PubMed]
- Gómez I, Seoane J, Varela-Centelles P, et al. Is diagnostic delay related to advanced-stage oral cancer? A meta-analysis. Eur J Oral Sci 2009;117:541-6. [Crossref] [PubMed]
- Onizawa K, Nishihara K, Yamagata K, et al. Factors associated with diagnostic delay of oral squamous cell carcinoma. Oral Oncol 2003;39:781-8. [Crossref] [PubMed]
- Jaulerry C, Bataini JP, Brunin F, et al. Prognostic factors and results of external irradiation of cancers of the base of the tongue. Ann Otolaryngol Chir Cervicofac 1985;102:519-24. [PubMed]
- Saito Y, Li L, Coyaud E, et al. LLGL2 rescues nutrient stress by promoting leucine uptake in ER(+) breast cancer. Nature 2019;569:275-9. [Crossref] [PubMed]
- Chen N, Jiang X, Wang J, et al. CXCL12-CXCR4/CXCR7 axis contributes to cell motilities of oral squamous cell carcinoma. Tumour Biol 2016;37:567-75. [Crossref] [PubMed]
- Ishikawa T, Nakashiro K, Klosek SK, et al. Hypoxia enhances CXCR4 expression by activating HIF-1 in oral squamous cell carcinoma. Oncol Rep 2009;21:707-12. [PubMed]
- Duan Y, Zhang S, Wang L, et al. Targeted silencing of CXCR4 inhibits epithelial-mesenchymal transition in oral squamous cell carcinoma. Oncol Lett 2016;12:2055-61. [Crossref] [PubMed]
- Faria PC, Sena AA, Nascimento R, et al. Expression of annexin A1 mRNA in peripheral blood from oral squamous cell carcinoma patients. Oral Oncol 2010;46:25-30. [Crossref] [PubMed]
- Nomura H, Uzawa K, Yamano Y, et al. Down-regulation of plasma membranous Annexin A1 protein expression in premalignant and malignant lesions of the oral cavity: correlation with epithelial differentiation. J Cancer Res Clin Oncol 2009;135:943-9. [Crossref] [PubMed]
- Lin CY, Jeng YM, Chou HY, et al. Nuclear localization of annexin A1 is a prognostic factor in oral squamous cell carcinoma. J Surg Oncol 2008;97:544-50. [Crossref] [PubMed]
- Zhang CH, Wang J, Zhang LX, et al. Shikonin reduces tamoxifen resistance through long non-coding RNA uc.57. Oncotarget 2017;8:88658-69. [Crossref] [PubMed]
- Wan YM, Tian J, Qi L, et al. ANXA1 affects cell proliferation, invasion and epithelial-mesenchymal transition of oral squamous cell carcinoma. Exp Ther Med 2017;14:5214-8. [Crossref] [PubMed]
- Rose AM, Krishan A, Chakarova CF, et al. MSR1 repeats modulate gene expression and affect risk of breast and prostate cancer. Ann Oncol 2018;29:1292-303. [Crossref] [PubMed]
- Seppälä EH, Ikonen T, Autio V, et al. Germ-line alterations in MSR1 gene and prostate cancer risk. Clin Cancer Res 2003;9:5252-6. [PubMed]
- Shiiba M, Saito K, Yamagami H, et al. Interleukin-1 receptor antagonist (IL1RN) is associated with suppression of early carcinogenic events in human oral malignancies. Int J Oncol 2015;46:1978-84. [Crossref] [PubMed]
- Shan J, Sun Z, Yang J, et al. Discovery and preclinical validation of proteomic biomarkers in saliva for early detection of oral squamous cell carcinomas. Oral Dis 2019;25:97-107. [Crossref] [PubMed]
- Leusink FK, van Diest PJ, Frank MH, et al. The Co-Expression of Kallikrein 5 and Kallikrein 7 Associates with Poor Survival in Non-HPV Oral Squamous-Cell Carcinoma. Pathobiology 2015;82:58-67. [Crossref] [PubMed]
- Zhao C, Zou H, Zhang J, et al. An integrated methylation and gene expression microarray analysis reveals significant prognostic biomarkers in oral squamous cell carcinoma. Oncol Rep 2018;40:2637-47. [Crossref] [PubMed]
- Srivastava RA. Regulation of the apolipoprotein E by dietary lipids occurs by transcriptional and post-transcriptional mechanisms. Mol Cell Biochem 1996;155:153-62. [Crossref] [PubMed]
- Pencheva N, Tran H, Buss C, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 2012;151:1068-82. [Crossref] [PubMed]
- Zheng P, Luo Q, Wang W, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis 2018;9:434. [Crossref] [PubMed]
- Jayakar SK, Loudig O, Brandwein-Gensler M, et al. Apolipoprotein E Promotes Invasion in Oral Squamous Cell Carcinoma. Am J Pathol 2017;187:2259-72. [Crossref] [PubMed]
- Pomerantz RG, Mirvish ED, Erdos G, et al. Novel approach to gene expression profiling in Sezary syndrome. Br J Dermatol 2010;163:1090-4. [Crossref] [PubMed]
- Shirai K, Shimada T, Yoshida H, et al. Interferon (IFN)-induced protein 35 (IFI35) negatively regulates IFN-beta-phosphorylated STAT1-RIG-I-CXCL10/CCL5 axis in U373MG astrocytoma cells treated with polyinosinic-polycytidylic acid. Brain Res 2017;1658:60-7. [Crossref] [PubMed]


