
Long non-coding RNAs regulating multiple proliferative pathways in cancer cell
Introduction
Long non-coding RNAs (lncRNAs) are implicated in the control of a series of cellular processes such as splicing, cell cycle, apoptosis, maintenance of pluripotency, and more. They basically exert this control by modulating the mechanisms of regulation of gene transcription and the translation of messenger RNAs, but also of protein localization within the cell and their degradation. They are able to take part in these mechanisms thanks to their ability to interact with a wide range of molecules including DNA, RNA and proteins of various type (1). The main functions that lncRNAs perform in the cell can be traced back to four basic categories: (I) molecular decoy, (II) molecular guide, (III) molecular scaffold and (IV) molecular regulator. As decoys, they can basically sequester microRNAs (miRNAs), thereby derepressing a series of molecular targets. As guides, they can guide multi-protein complexes able of inducing epigenetic changes to specific locations on the genome. As scaffolds, they participate directly in the formation of ribonucleoprotein complexes. Finally, as regulators, they can directly take part in the regulation of transcription (2). Given their plasticity of interaction and ability to act in different cellular mechanisms, it is not surprising that a vast repertoire of lncRNAs is deregulated in human carcinomas. In fact, their deregulated expression has been associated with different clinical-pathological characteristics such as survival, proliferation, the ability to metastasize, the immunological response and pluripotency, just to give examples.
The aim of this review, therefore, is to briefly describe the role of a small list of lncRNAs that are deregulated in human carcinomas and that control cell proliferation by acting at the same time on the main cell proliferative pathways. We have focused on a group of lncRNAs able to modulate proliferation of cancer cells either positively (H19, linc-ROR, MALAT1, HULC, HOTAIR and ANRIL), negatively (MEG3 and lincRNA-p21) and in a double fashion (UCA1 and TUG1).
Selected lncRNAs involved in cell proliferation
H19 imprinted maternally expressed transcript (H19)
The lncRNA H19, through the simultaneous action on different cellular pathways, undoubtedly represents one of the best examples of lncRNA regulating cell proliferation, in fact, its expression is essential for the development of various human cancers (3,4). Interestingly, H19 carries out its function of positive regulator of proliferation and tumorigenesis mainly through the interaction with EZH2 (5). This lncRNA is transcribed from the Igf2/H19 cluster, which also contains the IGF2 gene in addition to H19 (6). Interestingly, the H19 locus encodes a 2.9 kb lncRNA which also contains the miR-675 (7). H19 represents an example of imprinted gene, its expression being linked to the parental origin of the chromosome (8). A series of stimuli are able to activate the expression of lncRNA H19 and, among these, hypoxia, inflammation, but also growth factors and cytokines are certainly very important (9). The expression of H19 is also regulated by E2F1 (10). It is interesting to note that the silencing of H19 in pancreatic ductal adenocarcinoma cells induces a block of the cell cycle in the G0/G1 phase, as observed in a mouse model of carcinoma. The silencing of H19 also causes a reduction in the levels of E2F1 which, in turn, cannot longer induce and sustain the expression of H19, then blocking this regulatory positive loop (11).
Regulator of reprogramming (linc-ROR)
A series of studies report that linc-ROR is overexpressed in different human cancers and that its elevated levels are positively associated with a worse tumor prognosis (12). Interestingly, as far as cell proliferation is concerned, in pancreatic cancer stem cells (PCSCs) it has been observed that linc-ROR, by suppressing miR-145 and promoting the increase of Nanog, positively influences proliferation (13). In nasopharyngeal carcinoma, on the other hand, the function of linc-ROR is closely related to the control of the cell cycle, since its silencing induces an accumulation of cells in the G0/G1 phase accompanied by a decrease of those in the S phase (14). Among the factors that regulate the expression of linc-ROR, key factors regulating the maintenance of cellular pluripotency such as OCT4, SOX2 and Nanog, were reported to play a fundamental role (15).
Urothelial cancer associated 1 (UCA1)
LncRNA UCA1 plays an important role in the control of cancer cell proliferation. However, while in gastric carcinoma it is overexpressed acting as an oncogene (16), conversely, it is downregulated in esophageal squamous cell carcinoma blocking tumor growth, as demonstrated by functional experiments (17). It is interesting to note that UCA1, in addition to being induced by SP1 (18), is also induced by the presence of cancer associated fibroblast (CAF), as reported in colorectal cancer (19).
Maternally expressed gene 3 (MEG3)
In different types of cancer, MEG3 has been seen to perform its function as a tumor suppressor and negative regulator of cell proliferation, by targeting a series of miRNAs such as miR-421 in breast cancer (20), miR-181 in gastric cancer (21) and miR-21-5p in non-small cell lung cancer (NSCLC) (22). The methylation of its promoter undoubtedly represents one of the primary mechanisms by which MEG3 is regulated (23), however other factors are able to control its expression. In fact, cAMP is able to activate its expression by binding to a CRE element present in its promoter. It is interesting to note that the methylation state of the CRE element negatively affects the binding of cAMP, effectively blocking the activation of the promoter by this latter (24).
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
MALAT1 represents one of the basic regulator of cell cycle and proliferation, acting on these processes through different mechanisms. It has also been reported that this lncRNA is overexpressed in a series of human carcinomas (25), confirming its crucial role in cell proliferation. Interestingly, through the recruitment of SF2/ASF, MALAT1 can induce the transition of the cycle through the G0/G1 phase (26). Additionally, by translocating the hnRNP-C protein from the nucleus to the cytoplasm, MALAT1 is also able to control the transition of the cycle through the G2/M phase (27).
Taurine up-regulated 1 (TUG1)
TUG1 is an interesting lncRNA that can act as both tumor suppressor and oncogene, depending on the type of cancer in which it is deregulated. It acts on the regulation of cell proliferation mainly via two mechanisms: by sponge effect on miRNAs (28) and by interaction with the polycomb PRC1 and PRC2 complexes (29). As for the sponge effect mechanism on miRNAs, TUG1 performs this detrimental function on a long list of them. Interestingly, TUG1 positively regulates the expression of EZH2 through sponge effect on different miRNAs that target it, such as miR-144-3p (30) and miR-382 (31). Regarding its guide/scaffold function, it is interesting to note that in hepatocellular carcinoma TUG1 is able to target PRC2 on the KLF2 promoter (a tumor suppressor), repressing its expression (32).
Highly up-regulated in liver cancer (HULC)
LncRNA HULC has been found to be overexpressed in a wide range of human cancers such as hepatocellular, gastric, pancreatic and esophageal carcinoma (33). Interestingly, HULC is able to negatively regulate the expression of the tumor suppressor p18 which is located close to its position on the genome (34). It is worth to note that a study carried out in hepatocellular carcinoma shows that in the HULC promoter there is a binding site for CREB and that, therefore, its expression is controlled by this transcription factor (35).
HOX transcript antisense RNA (HOTAIR)
HOTAIR has been identified as a lncRNA transcribed by the HOXC cluster and able to carry out transcriptional repression on the HOXD cluster, by recruiting the PRC2 complex (36). A lot of studies showed that HOTAIR is responsible of the increased proliferation and progression along the cell cycle (among other things) in a wide range of human cancers. In particular, it has been observed that, through interaction with EZH2, HOTAIR promotes proliferation in glioma system (37). The interactions with the PRC2 complex, and more specifically with EZH2, represents one of the mechanisms by which HOTAIR acts. In fact, through this mechanism, the PRC2 complex is recalled on regions of the genome rich in GA by the interaction with HOTAIR, where it has previously been located (38). In this way, the PRC2 complex repositioned on the genome can perform its function as a repressor of gene expression by inducing the trimethylation of H3K27 (39,40). As far as HOTAIR regulation is concerned, it is worth noting how HOTAIR is regulated at the post-transcriptional level by miRNAs, such as miR-141 which displays tumor suppressor activity (41). NF-kB and estrogen receptor also seem to play an important role in the activation of HOTAIR, as reported in ovarian and breast cancer, respectively (42,43).
Tumor protein p53 pathway corepressor 1 (lincRNA-p21)
In general, lincRNA-p21 acts as a tumor suppressor lncRNA, negatively controlling proliferation and activating apoptosis (44). It is interesting to note that lincRNA-p21 acts itself as a transcriptional factor playing its critical role through the interaction with the transcriptional machinery (45). However, it has also been observed that lincRNA-p21 is able to regulate several functions by acting at post-transcriptional level (44).
CDKN2B antisense RNA 1 (ANRIL)
Like many other lncRNAs, ANRIL functions as an epigenetic transcriptional regulator of target genes. It is able to promote cell proliferation by directly acting on miRNAs and carrying out a sponge effect, as observed for miR-186 in cervical cancer (46) and miR-199a in breast cancer (47). However, ANRIL is also able to stimulate proliferation in prostate cancer cells by acting on the TGF-β1/SMAD pathway (48). E2F1 represents one of the most important activators of ANRIL, as it stimulates its promoter following genotoxic stress and DNA repair, when the cell can re-enter the cycle (49). Finally, it has also been reported that while SOX2 (50) and SP1 (51) positively regulate ANRIL, TET2 negatively regulates it (52).
Proliferative pathways regulated by lncRNAs
RB transcriptional corepressor 1 (pRB)
Several papers have reported that H19 is strongly involved in the pRB pathway (53,54). pRB is epigenetically regulated in different types of human cancer (55) and H19 participates in this kind of regulation through a mechanism involving miR-675 (which is contained in its sequence), targeting pRB and culminating in the promotion of cell proliferation. In fact, miR-675 is able to bind to the 3'-UTR of RB, negatively regulating its expression levels (53). Interestingly, in colorectal cancer and hepatocellular carcinoma it has been reported that the levels of H19/miR-675 and pRB levels are inversely correlated, thus confirming this regulatory mechanism (53,54). Therefore, H19/miR-675 are undoubtedly critical regulators of the pRB-induced tumor suppressor pathway. It is worth to note that pRB, in turn, through a repression of its promoter can suppress the expression of H19 induced by E2F1, revealing the existence of a self-regulatory mechanism between pRB and lncRNA H19 (Figure 1).
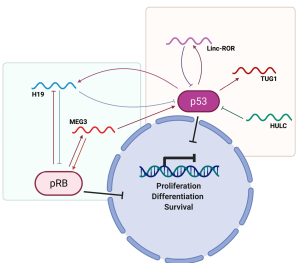
It is also interesting to note that MEG3 has the ability to trigger a decrease of cell proliferation through the MDM2 and pRB pathway, via the negative regulation of DNMT proteins and in particular DNMT3A (56). Decreased expression of MEG3 allows DNMT3A to further increase and repress MEG3 itself, giving rise to a self-regulatory feed-back loop. The pRB protein, on the other hand, is able to induce the expression of MEG3 (Figure 1) through the downregulation of DNMT1, with the consequent decrease of MEG3 methylation levels. In this way, MEG3 can increase its own expression and can regulate the proliferation of lung cells (57). It is also interesting to note that several miRNAs are able to activate the expression of MEG3 through the repression of DNMT1 and DNMT3b, as observed for miR-29a and miR-26a (58,59).
Tumor protein p53 (p53)
As observed in HCC, p53 activates the expression of H19 through the HIF-1 alpha pathway. H19, in turn, becomes part of a self-regulatory loop with p53 (Figure 1), since it can block its activity, preventing the expression of Bax and bypassing apoptosis, as reported in gastric cancer (60).
Similarly to what has been observed for H19, linc-ROR also appears to be both regulated and a regulator of p53 (Figure 1). In fact, if on the one hand p53 can induce its expression following cell damage (61), linc-ROR in turn is able to induce proliferation of colorectal cancer cells through the inhibition of p53 (62), by binding to the phosphorylated form of the hnRNP-I protein (61). Therefore, also in this case there is a reciprocal regulatory mechanism. However, it is interesting to note that only the wild type form of p53 has the ability to bind to the linc-ROR promoter and induce its activation (61).
Several studies have demonstrated that MEG3 carries out its activity through the action on the p53 pathway, then acting as a tumor suppressor negatively regulating cell proliferation. In colon carcinoma and in osteosarcoma cells, it was observed that MEG3 acts directly downregulating MDM2, thus promoting the activation of p53 (63). This mechanism involves the recruitment of the PRC2 complex on the MDM2 promoter by MEG3, emphasizing its role as a guide for the re-localization on the genome of epigenetic regulators of gene expression (63). Interestingly, in hepatoma cells it has also been observed that MEG3 directly interacts with the p53 protein, stabilizing it and prolonging its half-life. This mechanism triggers the downstream expression of several p53 targets which negatively regulate cell proliferation in different tumor histotypes (64).
In NSCLC and gliomas, TUG1 expression is strongly decreased compared to normal tissues (65). Thanks to a responsive element on its promoter, TUG1 is induced by p53 protein and can negatively regulate cell proliferation, exerting its function of tumor suppressor in different tumor systems.
It has also been reported that HULC is involved in p53 signaling, since it is able to block EEF1E, a critical activator of p53. This mechanism, subsequently, promotes tumor proliferation and growth (33).
Finally, lincRNA-p21, induced following DNA damage, can basically exert its function by inducing apoptosis through p53 (66). Interestingly, p53 itself is able to induce lincRNA-p21 expression by binding to its promoter. In particular, after exposure to UVB, lincRNA-p21 (induced through a p53-mediated mechanism) is responsible to trigger apoptotic death in UVB-treated keratinocytes (67). However, a mutation that hits one p53 allele, as early and frequently observed in skin cancer, is sufficient to confer an oncogenic function to p53, which is in turn reflected into an inhibitory effect on the lincRNA-p21 expression (67). Hence, the mutational status of p53 is crucial for lincRNA-p21 activation, and this occurrence is even more intricate than expected. In fact, in head and neck squamous cell carcinoma (HNSCC), it has been observed that also a mutant form of p53 is able to activate lincRNA-p21 expression, but this event directly relies on the involvement of nuclear transcription factor Y subunit alpha (NF-YA) in the transcriptional machinery (68).
Phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR)
H19 and miR-675 can activate the AKT/mTOR pathway and therefore cell proliferation, through the negative regulation of RUNX1 (69). In fact, H19 and miR-675 are overexpressed in gastric carcinoma and promote AKT phosphorylation, thereby stimulating the corresponding proliferative pathway (69). Additionally, in gallbladder cancer it has been observed that H19 exerts sponge effect on miR-194-5p, thus favoring the activation of AKT2 and strengthening cell proliferation (70). The PI3K/AKT pathway seems to be involved also in the regulation of H19, since in HCC it has been observed that TGF-β induces the expression of H19 through a mechanism that directly involves the PI3K protein (71), thus confirming a close relationship between this lncRNA and the PI3K/AKT pathway (Figure 2).
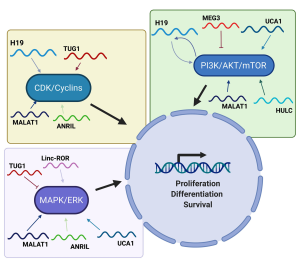
An interesting lncRNA able to positively regulate cell proliferation by activating the AKT pathway is undoubtedly UCA1. In gastric cancer, the expression levels of UCA1 are extremely high in tumor tissue compared to the normal counterpart. In this system, UCA1 directly interacts with EZH2 promoting its binding to the CCND1 promoter, to stimulate its transcription. In addition, UCA1 is also able to induce AKT phosphorylation. These two synergistic actions, thus, trigger the transition of cancer cells through the G1/S phase of the cell cycle (18). In bladder cancer, UCA1 influences the cell cycle through a mechanism that involves CREB, and which induces an increase in the expression levels of AKT1 and p-AKT1 (72). Furthermore, in bladder carcinoma cells, UCA1 also positively regulates glucose metabolism via the mTOR/STAT3/miR-143/hexokinase 2 pathway, taking part in the generation of the Warburg effect (73). Additionally, in NSCLC, it has been observed that UCA1 is overexpressed and regulates cell proliferation and drug resistance by directly activating the AKT/mTOR pathway (74).
Conversely, a series of studies has shown that MEG3 can inhibit the AKT pathway (Figure 2), then negatively regulating cell proliferation (75). While it is expressed in a series of normal tissues, MEG3 is decreased in a series of human cancers including glioma (76), meningioma (77), gastric (78) and breast cancer (75), letting to envisage its tumor suppressor role. This role is confirmed by functional experiments showing that the restored expression of MEG3 in breast and ovarian cancer cells results in a suppression of the PI3K/AKT pathway with a concomitant induction of PTEN expression (75,79). Interestingly, it has been reported that MEG3 exerts a repressive effect on cell proliferation, by blocking a series of miRNAs that target PTEN. In fact, in glioma cells, MEG3 negatively controls cell proliferation by blocking miR-19a, which in turn is able to target PTEN (80).
MALAT1 has been found overexpressed in a series of human cancers, then representing a clear example of oncogenic lncRNA. In particular, in gastric cancer it has been found overexpressed and functional studies have shown that this lncRNA induces the phosphorylation of AKT, PI3K and STAT3 (81,82). The ability of MALAT1 to induce proliferation by activating the PI3K/AKT pathway has also been observed in ovarian and cervical cancer (83,84).
In malignant melanoma, it has been reported that TUG1 can perform sponge effect on miR-129-5p. The decrease of this miRNA promotes the expression of the AEG1 protein, which plays a positive role on cell proliferation by activating the PI3K/AKT and WNT/β-catenin pathways (85). Interestingly, the PI3K/AKT pathway is also activated in osteosarcoma, where TUG1 is able to exert sponge effect on miR-219a-5p (86). Additionally, thanks to its ability to suppress HOXB7 expression, TUG1 is directly able of causing AKT dephosphorylation, with consequent pathway shutdown (65). Furthermore, TUG1 exerts sponge effect on several miRNAs targeting PTEN, then promoting its expression and further switching off the AKT pathway (87). Interestingly, the PI3K/AKT pathway in turn regulates the expression of TUG1 through an indirect mechanism. The expression of TUG1 is regulated by different transcription factors, among which it is worth mentioning SP1 (32), p53 (65) and FOXM1 (86). It is interesting to note that FOXM1 and TUG1 levels are positively correlated in osteosarcoma and that the former is able to directly transactivate the TUG1 promoter, following its activation by AKT (86).
The lncRNA HULC is abundantly expressed in pancreatic carcinoma where it stimulates cell proliferation by inhibiting miR-15a, with the consequent activation of the AKT pathway (88). Also in NSCLC, HULC is overexpressed and this is reflected in an increase of SPHK1 and in the downstream target PI3K/AKT pathway. HULC, therefore, induces the overexpression of SPHK1 and subsequent phosphorylation of AKT (89). In glioma cells, it has been demonstrated that the suppression of HULC induces a modulation of the PI3K/AKT/mTOR pathway (Figure 2) with a reduction of cell proliferation and block of cells in the G1/S phase (90). Interestingly, in glioma cells HULC is involved in the proliferation through the induction of ESM-1, then regulating the PI3K/AKT/mTOR pathway (90). As for the interaction with miRNAs, it has been observed that HULC is able to exert sponge effect on miR-122, that inactivates the PI3K/AKT, JAK/STAT and NOTCH pathways (91).
As far as HOTAIR is concerned, studies have revealed that, through its ability to recruit chromatin modifying complexes, it induces hypermethylation of the PTEN promoter with consequent reduction of its levels, then directly regulating the AKT pathway (92). In addition, HOTAIR is also able to address the recruitment of PCAF to a substantial list of genes involved in the PI3K/AKT/mTOR pathway (93).
As observed for MEG3, also lincRNA-p21 represses the PI3K/AKT pathway, then acting as tumor suppressor gene, as reported in prostate cancer, where its silencing is correlated with activation of the AKT protein (94). Moreover, as already observed for other lncRNAs, lincRNA-p21 exerts its functions through sponge effect on several miRNAs. It is worth noting that via sponge effect on miR-181b and miR-17-5p, it is able to modulate the AKT (promoting the re-expression of PTEN) and WNT (promoting the re-expression of WIF1, WNT inhibitory factor 1) activity, as reported in hepatic stellate cells (95,96).
Similarly, ANRIL promotes cell proliferation through the modulation of the PI3K/AKT pathway, as reported in osteosarcoma (97) and in glioma cells (98). In medulloblastoma, it was observed that ANRIL regulates the levels of phosphorylated p38 MAPK, ERK and AKT, with consequent effects on proliferation and apoptosis (99).
Finally, in NSCLC it has been reported that the expression of linc-ROR is positively associated with the levels of p-PI3K, p-AKT, and mTOR (100).
Cyclin-dependent kinases/cyclins/cyclin-dependent kinase inhibitors (CDKs/Cyclins/CDKi)
H19 is able to directly control crucial cell cycle regulators such as CCND1, CCNE1 and CDK4 (101), then stimulating the cell cycle through the sequestration of the eIF4A3 protein, the protein responsible for the splicing of pre-mRNAs.
MALAT1 is characterized by the ability to induce cell proliferation by controlling all phases of the cell cycle. In fact, following its experimental depletion, several positive regulators of the G0/G1 transition, such as CDC25 and cyclin A2, are no longer expressed. As far as G1 phase is concerned, it has been reported that MALAT1 depletion directly causes an increase of p53, as well as of p16, p21 and p27 (102). Finally, as regards the control of the G2/M phase, MALAT1 performs its function essentially through the control of B-MYB splicing (an oncogene involved in the G2/M transition), which in turn is responsible of the expression of several mitotic proteins, such as CDK1 and cyclin B1 (102). It is interesting to note that miR-101 and miR-217 can block MALAT1 in esophageal squamous cell carcinoma, then inducing a decrease in proliferation and an arrest of the cell cycle in the G2/M phase (103). This occurs because the lack of MALAT1 does not allow the increase of B-MYB and indeed promotes an increase of p21 and p27 levels (103).
By interacting with PRC2, TUG1 is able to induce epigenetic changes in the promoters of genes coding for CDK inhibitors, triggering their shutdown and promoting the advancement of cells along the cell cycle (Figure 2). Among these genes, it is worth mentioning p21, p15 and p16, p27 and p57 (104).
In nasopharyngeal carcinoma cells, it has been observed that the suppression of HULC induces an increase in the expression of p21 (105). Interestingly, it seems that the induction of p21 is indirect and depends on p53, which in turn is also induced by HULC. Namely, the overexpression of HULC in nasopharyngeal carcinoma induces cell growth by modulating the p53-p21 axis (105). Additionally, in liver carcinoma samples, the expression levels of HULC and p18 are inversely correlated, and this observation indicates that a close relationship between these two entities could exist. In fact, it is reported that HULC can negatively regulate the expression of p18 through a mechanism involving CREB. In particular, via CREB, the HBx protein activates HULC, which in turn represses the expression of p18 (34,35).
Interestingly, p21 is also subjected to the regulation by HOTAIR, as observed in lung adenocarcinoma, where HOTAIR promotes proliferation through its repression (106). However, HOTAIR can also regulate the expression of cyclins by acting as a decoy for several miRNAs with tumor suppressive functions. In ovarian cancer, in fact, HOTAIR targets miR-206, thus unblocking CCND1 and CCND2, which can thus carry out their positive action on the cell cycle (107). Again, in esophageal cancer HOTAIR is able to target miR-1, positively acting on the cell cycle control (108).
On the contrary, in an experimental mouse system it has been observed that lincRNA-p21 induces the expression of p21, reflecting its mechanism of action in the control of the G1/S transition and proliferation (45). In esophageal cancer cells, again, lincRNA-p21 is able to induce high levels of p21 and reduce cyclin D levels, causing cell cycle arrest (109). Additionally, in head and neck squamous carcinoma, it has been reported that lincRNA-p21 can also reduce the expression of cyclin B1 and cyclin D1, thus promoting an accumulation of cells in the G1 phase of the cell cycle (68).
It is worth to note that cyclins are also under the control of lncRNA UCA1. In fact, UCA1 induces the expression of cyclin D1 and stimulates the G1/S transition, interacting with EZH2 and promoting its binding to the cyclin D1 promoter. Indeed, UCA1 positively regulates cell proliferation through a mechanism that directly involves the interaction with EZH2.
ANRIL, like other lncRNAs, acts by regulating the expression of its target genes both through the interaction and recruitment of the PRC1 and PRC2 complexes, and through the sponge effect on several miRNAs (110). In particular, ANRIL through the recruitment of the PRC1 complex on the CDKN2A locus (through direct interaction with CBX7), is able to induce the epigenetic repression of p14ARF and p16INK4A (regulation in cis) with consequent effects on cell proliferation (111). Moreover, through interaction with SUZ12, ANRIL can recruit the PRC2 complex on the CDKN2B gene locus (regulation in cis), thus repressing the expression of p15INK4B (112). Interestingly, ANRIL by interacting with PRC2 can also silence KLF2 and p21 (regulation in trans), this resulting in an increased proliferation, as observed in NSCLC (113). In gastric cancer ANRIL promotes tumor growth through the transcriptional silencing of miR-99a and miR-449 via PRC2. Thanks to this mechanism, ANRIL has the ability to modulate the signaling downstream of mTOR and CDK6/E2F1 (114).
Mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK)
It has been reported that UCA1 is strongly expressed in lung cancer cell lines and that its silencing causes a block of cell proliferation. This silencing is directly reflected in a reduced expression of MAPK with lack of activation of the related pathway (115). It is interesting to note that the silencing of UCA1 leads directly to an overexpression of miR-143, which in turn causes suppression of MAPK (115).
Additionally, in ER-positive breast cancer, linc-ROR can stimulate proliferation by activating the MAPK/ERK (Figure 2) pathway through a mechanism that alters the stability of DUSP7, which in turn is able to downregulate ERK (116).
MALAT1 is involved in the proliferation of intestinal carcinoma cells, where it is abundantly expressed and exerts its function also modulating ERK/MAPK signaling. In fact, it has been observed that its depletion prevents the phosphorylation of MEK1/2, ERK1/2 and JNK (117).
As previously described lncRNAs, also TUG1 takes part in the regulation of MAPK/ERK pathway. Interestingly, it has been observed that following the silencing of TUG1, the levels of p-ERK, p-AKT and p-GSK3b increase and promote cell proliferation, underlining the involvement of TUG1 in the regulation of the AKT and MAPK pathways (65).
Finally, the involvement of ANRIL in the positive control of cell proliferation is also reflected in its ability to modulate the expression of p38, ERK and JNK, as observed in glioma cells (99).
Hypoxia-inducible factor 1 alpha (HIF-1 alpha) and hypoxia
A positive correlation between H19 and HIF-1 alpha expression in hypoxic tumor cells has been observed since several years (118). Furthermore, it has also been found that HIF-1 alpha is responsible to induce the expression of H19 by binding to its promoter and by induction of SP1 (119). Interestingly, H19 can regulate the expression of HIF-1 alpha through a mechanism based on the sponge effect exerted against miRNAs targeting HIF-1 alpha. In fact, in endometrial cancer, H19 up-regulates the levels of HIF-1 alpha (Figure 3) with consequent activation of the HIF-1 alpha/AXL signaling and increase in cell proliferation and tumor progression through the suppression of miR-20b-5p function (120).
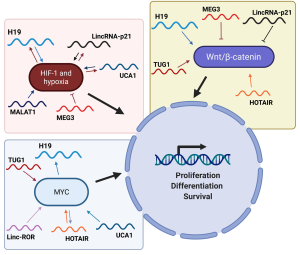
It is also worth to note that linc-ROR exerts sponge effect on miR-145, promoting an increase of HIF-1 alpha, which in turn induces VEGF, supporting cell proliferation (121).
UCA1 and HIF-1 alpha are linked to each other through a mechanism of mutual regulation (Figure 3). In fact, as observed in breast cancer (122), osteosarcoma (123) and bladder cancer (124), UCA1 is induced by high levels of HIF-1 alpha. UCA1, in turn, through sponge effect on miR-18a, is able to release HIF-1 alpha from the control of this miRNA, thus generating an interesting positive feed-back loop that plays a crucial role in hypoxic tumor cell proliferation.
An interesting work shows that MEG3 is involved in the regulation of HIF-1 alpha. In particular, it has been observed that after exposure of bronchial cells to nickel, an increased expression of DNMT3b represses the MEG3 promoter. This leads to a lack of transcription of PHLPP1, an AKT inhibitor, and the activation of the AKT/p70S64/S6/HIF-1 alpha pathway, with increased HIF-1 alpha expression and consequences on cell proliferation (125).
MALAT1 also appears to play a role in the control of proliferation in hypoxic conditions by acting on the HIF-1 alpha driven pathway. In fact, high levels of MALAT1 can induce stabilization of HIF-1 alpha, through blocking its ubiquitination, then promoting its accumulation (126). Furthermore, a study reported the role of MALAT1 as a hypoxia sensor (127), since it was observed that hypoxia directly induces MALAT1 expression in lung cancer cells. Finally, in multiple myeloma, it has been reported that HIF-1 alpha has the ability to induce MALAT1 via KDM3A, under hypoxic conditions. Since MALAT1, in turn, induces further accumulation of HIF-1 alpha (128), this mechanism generates a positive feed-back loop.
In hepatocellular carcinoma, hypoxia is associated with high levels of HOTAIR, that is able to induce HIF-1 alpha through a mechanism involving miR-130a-3p (129). Therefore, it is interesting to note that HIF-1 alpha is indeed a key target of HOTAIR. In renal carcinoma, a similar mechanism occurs, and this regulatory mechanism involves miR-127 which is blocked by HOTAIR through sponge effect. The induction of HIF-1 alpha, in turn, induces the AXL receptor and, thus, cell proliferation (130).
Finally, it has been shown that lincRNA-p21 and HIF-1 alpha are closely interconnected, letting to envisage that lincRNA-p21 is also involved in the signaling pathways activated under hypoxic conditions. In fact, while HIF-1 alpha can bind to lincRNA-p21 promoter activating it, lincRNA-p21, in turn, interferes with the VHL protein, promoting the stabilization of HIF-1 alpha, generating a positive feed-back loop (Figure 3) (131). However, in liver cells, lincRNA-p21 induces a decrease in the levels of HIF-1 alpha and, consequently of VEGF, thus blocking cell proliferation (132). Interestingly, in NSCLC it has been observed that lincRNA-p21 rather exhibits oncogenic behavior, as its high expression in some patient subtypes is associated with a poor outcome of the disease (133). In fact, under hypoxic conditions (when lincRNA-p21 is directly induced by HIF-1 alpha), lincRNA-p21 becomes a positive regulator of angiogenesis, rather than an inducer of apoptosis following the activation of p53 (131,133). Additionally, it is even more noteworthy that in NSCLC patients, lincRNA-p21 has been detected in the extracellular vesicles (EVs) present in the vessels that directly drain the lung tumor, and this occurrence is associated with a negative prognosis (134). In particular, the levels of lincRNA-p21 present in EVs are even higher in hypoxic conditions, and these levels directly trigger angiogenesis and alter tumor cells adhesion in target endothelial cells, confirming once again the role of lincRNA-p21 as an independent prognostic marker in NSCLC, and, in particular, in hypoxic conditions (134).
MYC proto-oncogene (MYC)
It is interesting to note that MYC can induce the transcription of H19 by binding to the conserved E-boxes present in its promoter (135). In particular, MYC induces histones acetylation, with a consequent activation of transcription. This is mainly important in leukemic cells where H19, induced by Bcr/Abl and MYC, promotes the tumorigenesis induced by Bcr/Abl through subsequent phosphorylation of STAT5 and expression of Bcl-Xl (136).
Linc-ROR, through the interaction with hnRNP-I, can strengthen the expression of MYC, contrary to what has been observed for p53 (137). In particular, the interaction with hnRNP-I promotes the binding of the protein to MYC mRNA, stabilizing it. Furthermore, it has also been observed that linc-ROR interacts with the AUF1 protein, thus preventing the latter from destabilizing the MYC messenger (137).
UCA1 carries out its positive effect on the regulation of proliferation also via action on the MYC pathway. In fact, high levels of UCA1 in multiple myeloma exert sponge effect on miR-331-3p which, if not blocked, would inhibit IL-6R. The latter activates MYC and the JAK2/STAT3 pathway, thus stimulating proliferation and inhibiting apoptosis (138).
One of the main mechanisms by which HOTAIR acts is characterized by the interaction with protein complexes capable of inducing epigenetic modifications in histones, and in particular PRC2 and LSD1 (139), thus emphasizing the role of HOTAIR as a scaffold. Indeed, in breast cancer cells, HOTAIR acts as scaffold for MYC, its cofactor HBHIP, and LSD1. The formation of this complex promotes the transcriptional activation of several molecular targets of MYC (such as cyclin A), which culminates in increased cell proliferation and oncogenesis (140). It is interesting to note that HOTAIR itself is under the control of MYC (Figure 3), this latter being able to directly bind to its promoter, as reported in gallbladder cancer (141).
Very interesting is the ability of TUG1 to act as a ceRNA for miR-145, the latter a potent tumor suppressor. In bladder cancer, in fact, it has been observed that these two non-coding RNAs regulate themselves in a reciprocal opposite way, becoming part of the RISC complex (142). Furthermore, in glioma stem cells it has been observed that TUG1 is able to block miR-145, thus preventing the degradation of SOX2 and MYC, which in turn support the stemness phenotype (143).
Wingless-type MMTV integration site family/β-catenin (WNT/β-catenin)
Like several other lncRNAs, H19 regulates cell proliferation via decoy effect on miRNAs. In particular, in colorectal cancer, H19 strengthens cell proliferation by exerting sponge effect on miR-200a, which in turn negatively regulates β-catenin (144).
Likewise, HOTAIR takes part in the modulation of proliferation by acting on the WNT/β-catenin pathway (Figure 3). In ductal pancreatic adenocarcinoma, treatment with radiotherapy increases the levels of HOTAIR, which in turn negatively regulates WNT inhibitory factor 1 (WIF-1), this latter being an inhibitor of the WNT pathway. Moreover, the experimental silencing of HOTAIR increases the levels of WIF-1 with consequent inhibition of the WNT pathway, a decrease in proliferation and an increase in radiotherapy sensitivity (145). HOTAIR can regulate the WNT/β-catenin pathway also by acting on miR-203a-3p, as reported in colorectal carcinoma cells, where HOTAIR promotes proliferation (146). In gastric cancer, on the other hand, HOTAIR has been reported to promote cell proliferation by decreasing miR-34a levels, with a consequent activation of the WNT pathway. Interestingly, in this system the levels of HOTAIR and miR-34a are inversely correlated (147).
Conversely, in NSCLC cells, MEG3 is able to block the cell cycle and activate apoptosis through p53 and the block of the WNT pathway. This occurrence is crucial as it allows to increase the drug sensitivity (148).
LincRNA-p21 directly regulates the WNT pathway (Figure 3), in colorectal cancer cells, by exerting a translational control on β-catenin (149). However, high levels of HuR inhibit lincRNA-p21 and promote translation of JUNB and CTNNB, which in turn promote cell proliferation (150). It is interesting to note that lincRNA-p21 performs its function by interacting with RNA binding proteins such as hnRNP-K and HuR. In particular, through the association with this latter and through the recruitment of the RISC complex, it controls the translation of β-catenin.
An interesting study in cervical cancer shows that TUG1 can neutralize miR-138-5p, with the consequent increase of the SIRT1 levels. In turn, SIRT1 induces the expression of several factors such as β-catenin, MYC and cyclin D1, thus resulting in the activation of the WNT/β-catenin pathway and induction of proliferation (151).
Finally, ANRIL, through the interaction with SOX2, has the ability to transcriptionally activate the WNT/β-catenin pathway (50). It is interesting to note that also linc-ROR and MALAT1 are able to influence the WNT/β-catenin pathway, as reported in ovarian cancer (152).
Conclusions
Thanks to the employ of new investigation technologies, much progress has recently been made in the scientific community with regard to the lncRNA research. As we have depicted in this review, only by considering a small number of them involved in the control of tumor cell proliferation, we can realize the vastness of the interactions and interconnections that are found within the molecular pathways of the cell. The scientific challenge of the future, therefore, will be to use -omics technologies to paint a clear picture of all the interactions involving non-coding RNAs, within each type of tumor. Undoubtedly, deciphering all these mechanisms of action will bring new possibilities for the treatment of human neoplastic diseases, especially as regards the implementation of novel therapeutic tools based on the use of lncRNAs.
Acknowledgments
The schematic representations reported in the figures were made with the support of Biorender Software (Biorender.com).
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Alfons Navarro, Joan Josep Castellano and Marina Díaz-Beyá) for the series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure forms (available at https://dx.doi.org/10.21037/tcr-21-230). The series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res 2018;3:108-17. [Crossref] [PubMed]
- Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577-87. [Crossref] [PubMed]
- Zhang DM, Lin ZY, Yang ZH, et al. IncRNA H19 promotes tongue squamous cell carcinoma progression through β-catenin/GSK3β/EMT signaling via association with EZH2. Am J Transl Res 2017;9:3474-86. [PubMed]
- Gabory A, Ripoche MA, Le Digarcher A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009;136:3413-21. [Crossref] [PubMed]
- Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659-65. [Crossref] [PubMed]
- Moon Y, Kim I, Chang S, et al. Hypoxia regulates allele-specific histone modification of the imprinted H19 gene. Biochim Biophys Acta Gene Regul Mech 2020;1863:194643. [Crossref] [PubMed]
- Lecerf C, Le Bourhis X, Adriaenssens E. The long non-coding RNA H19: an active player with multiple facets to sustain the hallmarks of cancer. Cell Mol Life Sci 2019;76:4673-87. [Crossref] [PubMed]
- Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005;280:29625-36. [Crossref] [PubMed]
- Ma L, Tian X, Wang F, et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther 2016;17:1051-61. [Crossref] [PubMed]
- Chen W, Yang J, Fang H, et al. Relevance function of linc-ROR in the pathogenesis of cancer. Front Cell Dev Biol 2020;8:696. [Crossref] [PubMed]
- Gao S, Wang P, Hua Y, et al. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget 2016;7:1608-18. [Crossref] [PubMed]
- Li L, Gu M, You B, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci 2016;107:1215-22. [Crossref] [PubMed]
- Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 2013;25:69-80. [Crossref] [PubMed]
- Wang CJ, Zhu CC, Xu J, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer 2019;18:115. Erratum in: Mol Cancer 2019;18:129. [Crossref] [PubMed]
- Wang X, Gao Z, Liao J, et al. lncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J Toxicol Environ Health A 2016;79:407-18. [Crossref] [PubMed]
- Wang ZQ, Cai Q, Hu L, et al. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis 2017;8:e2839. [Crossref] [PubMed]
- Jahangiri B, Khalaj-Kondori M, Asadollahi E, et al. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. J Cell Commun Signal 2019;13:53-64. [Crossref] [PubMed]
- Zhang W, Shi S, Jiang J, et al. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother 2017;91:312-9. [Crossref] [PubMed]
- Peng W, Si S, Zhang Q, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res 2015;34:79. [Crossref] [PubMed]
- Wang P, Chen D, Ma H, et al. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Onco Targets Ther 2017;10:5137-49. [Crossref] [PubMed]
- Moradi MT, Fallahi H, Rahimi Z. Interaction of long noncoding RNA MEG3 with miRNAs: A reciprocal regulation. J Cell Biochem 2019;120:3339-52. [Crossref] [PubMed]
- Zhao J, Zhang X, Zhou Y, et al. Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int J Biochem Cell Biol 2006;38:1808-20. [Crossref] [PubMed]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9:703-19. [Crossref] [PubMed]
- Wang J, Su L, Chen X, et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother 2014;68:557-64. [Crossref] [PubMed]
- Yang F, Yi F, Han X, et al. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett 2013;587:3175-81. [Crossref] [PubMed]
- Li J, Zhang Q, Fan X, et al. The long noncoding RNA TUG1 acts as a competing endogenous RNA to regulate the Hedgehog pathway by targeting miR-132 in hepatocellular carcinoma. Oncotarget 2017;8:65932-45. [Crossref] [PubMed]
- Zhou H, Sun L, Wan F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol Lett 2019;18:4393-402. [Crossref] [PubMed]
- Cao J, Han X, Qi X, et al. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol 2017;51:1115-23. [Crossref] [PubMed]
- Zhao L, Sun H, Kong H, et al. The lncRNA-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging mir-382. Cell Physiol Biochem 2017;42:2145-58. [Crossref] [PubMed]
- Huang MD, Chen WM, Qi FZ, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer 2015;14:165. [Crossref] [PubMed]
- Yu X, Zheng H, Chan MT, et al. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med 2017;21:410-7. [Crossref] [PubMed]
- Du Y, Kong G, You X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 2012;287:26302-11. [Crossref] [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Ghafouri-Fard S, Dashti S, Farsi M, et al. HOX transcript antisense RNA: an oncogenic lncRNA in diverse malignancies. Exp Mol Pathol 2021;118:104578. [Crossref] [PubMed]
- Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015;6:537-46. [Crossref] [PubMed]
- Chu C, Qu K, Zhong FL, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011;44:667-78. [Crossref] [PubMed]
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 2004;18:1592-605. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Chiyomaru T, Fukuhara S, Saini S, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem 2014;289:12550-65. [Crossref] [PubMed]
- Özeş AR, Miller DF, Özeş ON, et al. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016;35:5350-61. [Crossref] [PubMed]
- Bhan A, Mandal SS. Estradiol-induced transcriptional regulation of long non-coding RNA, HOTAIR. Methods Mol Biol 2016;1366:395-412. [Crossref] [PubMed]
- Tang SS, Zheng BY, Xiong XD. LincRNA-p21: implications in human diseases. Int J Mol Sci 2015;16:18732-40. [Crossref] [PubMed]
- Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014;54:777-90. [Crossref] [PubMed]
- Zhang JJ, Wang DD, Du CX, et al. Long Noncoding RNA ANRIL Promotes Cervical Cancer Development by Acting as a Sponge of miR-186. Oncol Res 2018;26:345-52. [Crossref] [PubMed]
- Xu ST, Xu JH, Zheng ZR, et al. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in triple-negative breast cancer. Biomed Pharmacother 2017;96:14-21. [Crossref] [PubMed]
- Zhao B, Lu YL, Yang Y, et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-beta1/Smad signaling pathway. Cancer Biomark 2018;21:613-20. [Crossref] [PubMed]
- Wan G, Mathur R, Hu X, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal 2013;25:1086-95. [Crossref] [PubMed]
- Wu JH, Tang JM, Li J, et al. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci Rep 2018;8:3333. [Crossref] [PubMed]
- Huang MD, Chen WM, Qi FZ, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol 2015;8:50. [Crossref] [PubMed]
- Deng W, Wang J, Zhang J, et al. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life 2016;68:355-64. [Crossref] [PubMed]
- Tsang WP, Ng EK, Ng SS, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010;31:350-8. [Crossref] [PubMed]
- Hernandez JM, Elahi A, Clark CW, et al. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann Surg Oncol 2013;20:S625-35. [Crossref] [PubMed]
- Ohtani-Fujita N, Dryja TP, Rapaport JM, et al. Hypermethylation in the retinoblastoma gene is associated with unilateral, sporadic retinoblastoma. Cancer Genet Cytogenet 1997;98:43-9. [Crossref] [PubMed]
- Lyu Y, Lou J, Yang Y, et al. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia 2017;31:2543-51. [Crossref] [PubMed]
- Kruer TL, Dougherty SM, Reynolds L, et al. Expression of the lncRNA maternally expressed gene 3 (MEG3) contributes to the control of lung cancer cell proliferation by the Rb pathway. PLoS One 2016;11:e0166363. [Crossref] [PubMed]
- Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011;30:4750-6. [Crossref] [PubMed]
- Jia LF, Wei SB, Gan YH, et al. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer 2014;135:2282-93. [Crossref] [PubMed]
- Zhang L, Zhou Y, Huang T, et al. The interplay of lncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci 2017;18:450. [Crossref] [PubMed]
- Zhang A, Zhou N, Huang J, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res 2013;23:340-50. [Crossref] [PubMed]
- Li H, Jiang X, Niu X. Long non-coding RNA reprogramming (ROR) promotes cell proliferation in colorectal cancer via affecting P53. Med Sci Monit 2017;23:919-28. [Crossref] [PubMed]
- Zhou Y, Zhong Y, Wang Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 2007;282:24731-42. [Crossref] [PubMed]
- Zhu J, Liu S, Ye F, et al. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One 2015;10:e0139790. [Crossref] [PubMed]
- Zhang EB, Yin DD, Sun M, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 2014;5:e1243. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Hall JR, Messenger ZJ, Tam HW, et al. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis 2015;6:e1700. [Crossref] [PubMed]
- Jin S, Yang X, Li J, et al. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol Cancer 2019;18:38. [Crossref] [PubMed]
- Liu G, Xiang T, Wu QF, et al. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells. Oncol Res 2016;23:99-107. [Crossref] [PubMed]
- Wang SH, Wu XC, Zhang MD, et al. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumour Biol 2016;37:9721-30. [Crossref] [PubMed]
- Matouk IJ, Raveh E, Abu-lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 2014;1843:1414-26. [Crossref] [PubMed]
- Yang C, Li X, Wang Y, et al. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene 2012;496:8-16. [Crossref] [PubMed]
- Li Z, Li X, Wu S, et al. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci 2014;105:951-5. [Crossref] [PubMed]
- Cheng N, Cai W, Ren S, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015;6:23582-93. [Crossref] [PubMed]
- Zhang CY, Yu MS, Li X, et al. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol 2017;39:1010428317701311. [Crossref] [PubMed]
- Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 2012;113:1868-74. [Crossref] [PubMed]
- Zhang X, Gejman R, Mahta A, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 2010;70:2350-8. [Crossref] [PubMed]
- Sun M, Xia R, Jin F, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol 2014;35:1065-73. [Crossref] [PubMed]
- Wang J, Xu W, He Y, et al. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res 2018;67:927-36. [Crossref] [PubMed]
- Qin N, Tong GF, Sun LW, et al. Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol Res 2017;25:1471-8. [Crossref] [PubMed]
- Zhu K, Ren Q, Zhao Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett 2019;17:5335-42. [Crossref] [PubMed]
- Dai Q, Zhang T, Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag Res 2020;12:1929-39. [Crossref] [PubMed]
- Jin Y, Feng SJ, Qiu S, et al. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2017;21:3176-84. [PubMed]
- Zhao F, Fang T, Liu H, et al. Long non-coding RNA MALAT1 promotes cell proliferation, migration and invasion in cervical cancer by targeting miR-625-5p and AKT2. Panminerva Med 2020; Epub ahead of print. [Crossref] [PubMed]
- Long J, Menggen Q, Wuren Q, et al. Long noncoding RNA taurine-upregulated gene1 (TUG1) promotes tumor growth and metastasis through TUG1/mir-129-5p/astrocyte-elevated gene-1 (AEG-1) axis in malignant melanoma. Med Sci Monit 2018;24:1547-59. [Crossref] [PubMed]
- Li Y, Zhang T, Zhang Y, et al. Targeting the FOXM1-regulated long noncoding RNA TUG1 in osteosarcoma. Cancer Sci 2018;109:3093-104. [Crossref] [PubMed]
- Li J, An G, Zhang M, et al. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem Biophys Res Commun 2016;477:743-8. [Crossref] [PubMed]
- Feng H, Wei B, Zhang Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int J Biol Macromol 2019;126:891-8. [Crossref] [PubMed]
- Liu L, Zhou XY, Zhang JQ, et al. LncRNA HULC promotes non-small cell lung cancer cell proliferation and inhibits the apoptosis by up-regulating sphingosine kinase 1 (SPHK1) and its downstream PI3K/Akt pathway. Eur Rev Med Pharmacol Sci 2018;22:8722-30. [PubMed]
- Zhu Y, Zhang X, Qi L, et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget 2016;7:14429-40. [Crossref] [PubMed]
- Kong D, Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J Cell Biochem 2018;119:1050-61. Retraction in: J Cell Biochem 2021;122:599. [Crossref] [PubMed]
- Li D, Feng J, Wu T, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol 2013;182:64-70. [Crossref] [PubMed]
- Hatziapostolou M, Koutsioumpa M, Kottakis F, et al. Targeting colon cancer metabolism through a long noncoding RNA. J Clin Oncol 2015;33:604. [Crossref]
- Wang X, Xu Y, Jiang C, et al. LincRNA-p21 suppresses development of human prostate cancer through inhibition of PKM2. Cell Prolif 2017;50:e12395. [Crossref] [PubMed]
- Yu F, Lu Z, Chen B, et al. Identification of a novel lincRNA-p21-miR-181b-PTEN signaling cascade in liver fibrosis. Mediators Inflamm 2016;2016:9856538. [Crossref] [PubMed]
- Yu F, Guo Y, Chen B, et al. LincRNA-p21 inhibits the Wnt/beta-catenin pathway in activated hepatic stellate cells via sponging microRNA-17-5p. Cell Physiol Biochem 2017;41:1970-80. [Crossref] [PubMed]
- Yu G, Liu G, Yuan D, et al. Long non-coding RNA ANRIL is associated with a poor prognosis of osteosarcoma and promotes tumorigenesis via PI3K/Akt pathway. J Bone Oncol 2018;11:51-5. [Crossref] [PubMed]
- Dong X, Jin Z, Chen Y, et al. Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem 2018;119:2708-18. [Crossref] [PubMed]
- Zhang H, Wang X, Chen X. Potential role of long non-coding RNA ANRIL in pediatric medulloblastoma through promotion on proliferation and migration by targeting miR-323. J Cell Biochem 2017;118:4735-44. [Crossref] [PubMed]
- Shi H, Pu J, Zhou XL, et al. Silencing long non-coding RNA ROR improves sensitivity of non-small-cell lung cancer to cisplatin resistance by inhibiting PI3K/Akt/mTOR signaling pathway. Tumour Biol 2017;39:1010428317697568. [Crossref] [PubMed]
- Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 2016;7:22159-73. [Crossref] [PubMed]
- Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368. [Crossref] [PubMed]
- Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem 2015;290:3925-35. [Crossref] [PubMed]
- Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis 2016;7:e2109. [Crossref] [PubMed]
- Jiang X, Liu W. Long Noncoding RNA Highly Upregulated in Liver Cancer Activates p53-p21 Pathway and Promotes Nasopharyngeal Carcinoma Cell Growth. DNA Cell Biol 2017;36:596-602. [Crossref] [PubMed]
- Liu Y, Wang B, Liu X, et al. Epigenetic silencing of p21 by long non-coding RNA HOTAIR is involved in the cell cycle disorder induced by cigarette smoke extract. Toxicol Lett 2016;240:60-7. [Crossref] [PubMed]
- Chang L, Guo R, Yuan Z, et al. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem 2018;49:1289-303. [Crossref] [PubMed]
- Ren K, Li Y, Lu H, et al. Long noncoding RNA HOTAIR controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. Transl Oncol 2016;9:489-97. [Crossref] [PubMed]
- Zhang Y, Miao Y, Shang M, et al. LincRNA-p21 leads to G1 arrest by p53 pathway in esophageal squamous cell carcinoma. Cancer Manag Res 2019;11:6201-14. [Crossref] [PubMed]
- Kong Y, Hsieh CH, Alonso LC. ANRIL: A lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne) 2018;9:405. [Crossref] [PubMed]
- Yap KL, Li S, Munoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662-74. [Crossref] [PubMed]
- Kotake Y, Nakagawa T, Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011;30:1956-62. [Crossref] [PubMed]
- Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 2015;14:268-77. [Crossref] [PubMed]
- Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 2014;5:2276-92. [Crossref] [PubMed]
- Jun T, Zheng FS, Ren KM, et al. Long non-coding RNA UCA1 regulates the proliferation, migration and invasion of human lung cancer cells by modulating the expression of microRNA-143. Eur Rev Med Pharmacol Sci 2018;22:8343-52. [PubMed]
- Peng WX, Huang JG, Yang L, et al. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol Cancer 2017;16:161. [Crossref] [PubMed]
- Wu XS, Wang XA, Wu WG, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther 2014;15:806-14. [Crossref] [PubMed]
- Matouk IJ, Mezan S, Mizrahi A, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta 2010;1803:443-51. [Crossref] [PubMed]
- Wu W, Hu Q, Nie E, et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep 2017;7:45029. [Crossref] [PubMed]
- Zhu H, Jin YM, Lyu XM, et al. Long noncoding RNA H19 regulates HIF-1alpha/AXL signaling through inhibiting miR-20b-5p in endometrial cancer. Cell Cycle 2019;18:2454-64. [Crossref] [PubMed]
- Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem 2016;40:219-29. [Crossref] [PubMed]
- Li X, Wu Y, Liu A, et al. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1alpha feedback regulatory loop. Tumour Biol 2016;37:14733-43. [Crossref] [PubMed]
- Li T, Xiao Y, Huang T. HIF1alphainduced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol Rep 2018;39:1072-80. [PubMed]
- Xue M, Li X, Li Z, et al. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol 2014;35:6901-12. [Crossref] [PubMed]
- Zhou C, Huang C, Wang J, et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1alpha translation. Oncogene 2017;36:3878-89. [Crossref] [PubMed]
- Luo F, Liu X, Ling M, et al. The lncRNA MALAT1, acting through HIF-1alpha stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta 2016;1862:1685-95. [Crossref] [PubMed]
- Hu L, Tang J, Huang X, et al. Hypoxia exposure upregulates MALAT-1 and regulates the transcriptional activity of PTB-associated splicing factor in A549 lung adenocarcinoma cells. Oncol Lett 2018;16:294-300. [Crossref] [PubMed]
- Ikeda S, Kitadate A, Abe F, et al. Hypoxia-inducible KDM3A addiction in multiple myeloma. Blood Adv 2018;2:323-34. [Crossref] [PubMed]
- Hu M, Fu Q, Jing C, et al. LncRNA HOTAIR knockdown inhibits glycolysis by regulating miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. Biomed Pharmacother 2020;125:109703. [Crossref] [PubMed]
- Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis 2017;8:e2772. [Crossref] [PubMed]
- Yang F, Zhang H, Mei Y, et al. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 2014;53:88-100. [Crossref] [PubMed]
- Ye Y, Peng Y, Li Y, et al. Effect of lincRNA-p21 targeting HIF-1alpha on biological functions of liver cancer cells. Oncol Lett 2019;17:4964-8. [PubMed]
- Castellano JJ, Navarro A, Vinõlas N, et al. LincRNA-p21 impacts prognosis in resected non-small cell lung cancer patients through angiogenesis regulation. J Thorac Oncol 2016;11:2173-82. [Crossref] [PubMed]
- Castellano JJ, Marrades RM, Molins L, et al. Extracellular vesicle lincRNA-p21 expression in tumor-draining pulmonary vein defines prognosis in NSCLC and modulates endothelial cell behavior. Cancers (Basel) 2020;12:734. [Crossref] [PubMed]
- Barsyte-Lovejoy D, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006;66:5330-7. [Crossref] [PubMed]
- Guo G, Kang Q, Chen Q, et al. High expression of long non-coding RNA H19 is required for efficient tumorigenesis induced by Bcr-Abl oncogene. FEBS Lett 2014;588:1780-6. [Crossref] [PubMed]
- Huang J, Zhang A, Ho TT, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res 2016;44:3059-69. [Crossref] [PubMed]
- Li JL, Liu XL, Guo SF, et al. Long noncoding RNA UCA1 regulates proliferation and apoptosis in multiple myeloma by targeting miR-331-3p/IL6R axis for the activation of JAK2/STAT3 pathway. Eur Rev Med Pharmacol Sci 2019;23:9238-50. [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689-93. [Crossref] [PubMed]
- Li Y, Wang Z, Shi H, et al. HBXIP and LSD1 scaffolded by lncRNA Hotair mediate transcriptional activation by c-Myc. Cancer Res 2016;76:293-304. [Crossref] [PubMed]
- Ma MZ, Li CX, Zhang Y, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer 2014;13:156. [Crossref] [PubMed]
- Tan J, Qiu K, Li M, et al. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett 2015;589:3175-81. [Crossref] [PubMed]
- Katsushima K, Natsume A, Ohka F, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun 2016;7:13616. [Crossref] [PubMed]
- Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing beta-catenin expression in colorectal cancer. Biomed Res Int 2017;2017:2767484. [PubMed]
- Jiang Y, Li Z, Zheng S, et al. The long non-coding RNA HOTAIR affects the radiosensitivity of pancreatic ductal adenocarcinoma by regulating the expression of Wnt inhibitory factor 1. Tumour Biol 2016;37:3957-67. [Crossref] [PubMed]
- Xiao Z, Qu Z, Chen Z, et al. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via miR-203a-3p-mediated Wnt/ss-catenin signaling pathway. Cell Physiol Biochem 2018;46:1275-85. [Crossref] [PubMed]
- Cheng C, Qin Y, Zhi Q, et al. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol 2018;107:2620-9. [Crossref] [PubMed]
- Xia Y, He Z, Liu B, et al. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/beta-catenin signaling pathway. Mol Med Rep 2015;12:4530-7. [Crossref] [PubMed]
- Wang G, Li Z, Zhao Q, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol Rep 2014;31:1839-45. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012;47:648-55. [Crossref] [PubMed]
- Zhu J, Shi H, Liu H, et al. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget 2017;8:65253-64. [Crossref] [PubMed]
- Lou Y, Jiang H, Cui Z, et al. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/beta-catenin signaling. Oncotarget 2017;8:69983-94. [Crossref] [PubMed]


