
Genome-wide expression changes mediated by A-to-I RNA editing correlate with hepatic oncogenesis
Introduction
Adenosine-to-inosine RNA editing, abbreviated as A-to-I RNA editing, is a highly conserved type of RNA modification widespread across the animal kingdom (1,2). The editing enzyme is known as adenosine deaminase acting on RNA (ADAR) (3). Unlike other RNA modifications such as methylation (4), the A-to-I editing process is simply controlled by ADARs and no other “readers” (5) or “erasers” (6). This largely facilitated the study of editing regulation because no other causal factors would confound the “ADAR-editing events’ relationship. Nevertheless, different clades have different numbers of ADAR proteins. Mammals such as humans bear three ADAR proteins: ADAR1 (gene ADAR), ADAR2 (gene ADARB1), and ADAR3 (gene ADARB2) (7), whereas in arthropods there is only one ADAR protein homologous to mammalian ADAR2 (8,9). In other taxa, one to three ADAR proteins have been identified (3).
Due to the preference of the editing enzyme, A-to-I editing events do not randomly occur on RNAs; instead, they are more likely to take place on adenosines within specific regions such as double-stranded RNA structures (8,10). The genomic locations of editing sites in many species have been recorded in databases and in the literature (11,12). For instance, mammalian editing sites are mainly located in repetitive transposable elements. The total number of editing sites varies drastically among different species (2,13,14). In humans, millions of adenosine sites are considered editable (15). Therefore, to study the editome in a new sample, a simple method is to directly retrieve the editing status of known editable sites (16), which saves much effort in dealing with variant calling.
Apart from the collection and annotation of editing sites, the most important task is to predict the function of these editing events. To date, A-to-I editing has been shown to affect many biological processes from multiple aspects. Most of the biological functions of A-to-I editing rely on the fact that inosines are interpreted as guanosines in the cellular environment. In other words, A-to-I editing produces a consequence similar to A-to-G DNA mutation. However, one difference is that A-to-G DNA mutations are permanent, while A-to-I RNA editing is controllable. This allows editing to selectively take place at different times and places in a flexible manner, which facilitates an organism’s ability to switch between different alleles when necessary.
Based on the A-to-G property of A-to-I editing, the most obvious consequence is that editing could change the amino acid sequence of protein-coding regions. Therefore, editing events in coding sequence (CDS) are usually subjected to strong natural selection (17-20). Editing events in non-coding regions are not always silent. A-to-I events involving RNA splicing regions usually alter the splicing patterns of pre-mRNAs (21,22), and the altered RNA structure may affect the editing efficiency of other editable sites (23). Splicing patterns usually determine the length of mature mRNAs (24), which is an important factor correlated with the expression and translation levels of host genes (25). Moreover, the A-to-I switch in 3’-UTRs could cause the gain or loss of microRNA binding sites, and editing in microRNA seed regions could affect target selection by microRNAs (26). As a consequence, A-to-I editing sites related to microRNA networks can also modify the expression and translation levels of target genes.
With the introduction of multiple functions of A-to-I RNA editing, there is no reason to omit the role of RNA editing in many human diseases. In particular, in the cancer field, the rise of single-cell sequencing techniques has caused many researchers to focus only on gene expression changes between tumor and normal samples. However, when one talks about gene expression, what is the definition of “a gene”? One gene has different isoforms (transcripts), translation start and stop sites (27,28), allele-specific expression caused by mutations (29,30), and one isoform could have different combinations of RNA editing events, and these molecules are actually different alleles (with potentially different functions), but they are usually pooled and regarded as the same “gene.” The automatic omission of RNA editing information is arbitrary and may miss important clues. For example, it has been reported that A-to-I editing-mediated isoform switches could lead to leukemia (31). Moreover, many human diseases are associated with RNA editing (32). This reminds us that many messages might be lost if one focuses solely on gene expression in cancer studies: the focus on cancer studies needs to be redirected. Different RNA modifications and isoform switches cannot be captured by single-cell sequencing; therefore, only traditional sequencing methods could retrieve this information.
In this study, we fully take advantage of the transcriptomes and translatomes of liver cancer and normal tissues from ten patients. We profiled the landscape of the A-to-I RNA editome in these samples (33). Globally, editing activity was enhanced in all tumor samples compared to that in normal samples. Expression of the gene encoding the RNA editing enzyme ADAR was also elevated. Two intronic self-editing sites in ADAR mRNA controlled the splicing pattern of ADAR and might regulate its translation efficiency (TE). Moreover, the expression of oncogenes is generally upregulated in tumors, whereas tumor suppressor genes (TSG) were downregulated, possibly due to alterations to microRNA binding regions or RNA splicing sites caused by A-to-I editing.
In conclusion, we demonstrated that A-to-I RNA editing plays a crucial role in the oncogenesis of liver cancer. ADAR regulates its own expression by self-editing, and it also affects the global transcriptomes and translatomes of cancer-related genes by editing them and changing their expression patterns. Again, we emphasize that RNA modifications should not be ignored in human disease studies, including in cancer research.
We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/tcr-21-236).
Methods
Data collection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Human reference genomes were downloaded from the Ensembl website (http://ensemblgenomes.org/). Sequencing data (RNA-Seq and ribosome profiling) of normal/tumor tissues from ten patients were downloaded from NCBI (accession number GSE112705) (33). According to the original literature, the ten patients had hepatocellular carcinoma (HCC). Data involving liver tumor tissues and adjacent normal liver tissues were collected.
Lists of oncogenes and TSGs were downloaded from the Cancer Gene Consortium website (CGC, https://cancer.sanger.ac.uk/census/). A total of 240 oncogenes (Table S1) and 242 TSGs (Table S2) were identified. This final gene list did not include ambiguously annotated genes (Supplementary file: Table S3); for example, some genes were annotated as both oncogenes and TSGs. These genes were not included in the list.
Mapping and variant calling
Sequencing reads were mapped to the human reference genome (build hg38) using STAR (34) with default parameters (v.2.7.3a). Three mismatches were allowed for each read in the alignment. Reads spanning RNA splicing junctions were split into two parts that were then each aligned to different genomic locations. Multiple mappers were allowed so that each read could be mapped to multiple locations in the genome. However, in these samples, 79–96% of the input reads were unique mappers that mapped to a single location in the genome. Only unique mappers were used in subsequent analyses. Gene expression profiles were downloaded according to an original article (33). Variants were called using the software Samtools (35). Default parameters were used except for base quality >25 and mapping quality >20. The editing ratio at each site was defined as the edited allele (G allele) divided by the total coverage at each site. There were ten patients each with RNA-Seq and ribo-seq data for tumor and normal samples, and a total of 40 samples were analyzed. In each sample, we selected sites with coverage ≥5, edited allele count (G allele) ≥2, and editing ratio ≥0.1. The genomic coordinates of these sites were combined (union set) to obtain a list of high-confidence editing sites for these 40 samples. These sites were anticipated to be bona fide editing sites because the criteria ensured adequate coverage of G alleles that were unlikely to derive from sequencing errors (since the sequencing error rate was <1%).
A total of 5,061 high-confidence sites were identified. Functional annotation of editing sites was accomplished by locating the editing site coordinates with reference to the genome annotation file because the genome annotation file contains the genomic coordinates of all genes, exons, and coding regions.
TE
A general measurement of TE is the ribosome occupancy calculated from mRNA-Seq and ribosome profiling data. The sequenced short reads in the ribosome profiling data were termed ribosome-protected fragments (RPF). The TE of genes was calculated as RPKMRPF/RPKMmRNA. (RPKM = “reads per kilobase per million mapped reads.” The raw read count of each gene was determined using the software package htseq (36).
Statistical analysis
Graphic works and statistics were obtained in the R environment (v.3.5.2). Boxplots were plotted using a command “boxplot()”. Barplots were plotted using a command “barplot()”. Dotplots were plotted using a command “plot()”. Heatmaps were created by package “pheatmap”. KS (Kolmogorov-Smirnov) tests were performed using the command “ks.test()”. The Spearman correlation coefficient between two vectors was calculated using the function cor.test (x, y, method = ‘spearman’).
Results
Calling high-confidence editing sites in normal and tumor samples
We mapped transcriptome (RNA-Seq) and translatome (ribosome profiling, ribo-seq) reads to the human reference genome (build hg38). We called the variants in the next generation sequencing (NGS) data and extracted the read count for known human A-to-I editing sites (12). Next, it was necessary to discard the unreliable variants and retain the high-confidence editing sites. There were ten patients each with RNA-Seq and ribo-seq data for tumor and normal samples, and a total of 40 samples were analyzed. In each sample, we selected sites with coverage ≥5 and edited allele count (G allele) ≥2. The genomic coordinates of these sites were combined (union set) to obtain a list of editing sites in these 40 samples.
A total of 5,061 unique editing sites were identified. Of these, 3,082 were located in the 3'-UTRs of genes, 733 sites were located in introns, 77 in 5'-UTRs, 115 caused missense mutations, and 49 caused synonymous mutations (Figure 1A,B). Generally, editing sites in coding regions comprised only a very small proportion of the total editing sites. In accordance with known facts, we found that 4,521 (~90%) of our editing sites were located in Alu repeats (Figure 1C), and the fraction of sites in 3’-UTRs was higher for Alu sites (63%) than for non-Alu sites (41%) (Figure 1D).
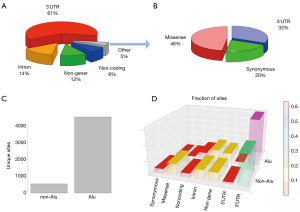
As stated, these 5,061 unique sites represent the total list of editing sites. Since there were four samples (RNA/ribo, normal/tumor) for each patient, we needed to parse the editing status involving these 5,061 sites for each sample.
Globally elevated editing efficiency in tumor samples
For exploratory RNA editing comparison, we first briefly tested for correlations among 10 tumor samples and 10 normal samples. Our goal was to determine whether tumor samples were more variable than normal samples. RNA-Seq and riboseq were investigated separately. Regardless of gene expression (RPKM, Figure 2A,B) or A-to-I editing ratios (Figure 2C,D), there were clear trends that (I) normal samples had greater pairwise correlation with each other; (II) tumor samples exhibited lower pairwise correlations with each other; (III) the correlation between normal and tumor samples was lowest. A simple explanation for this observation was that tumor samples were more variable due to the unpredictability of oncogenesis and tumor development, leading to fluctuations in gene expression or RNA editing spectra in tumor samples.

Next, we calculated the mean editing ratio for each sample. Strikingly, for RNA-Seq data, editing ratios were higher in tumors for all tumor-normal pairs (Figure 3A). For ribo-seq data, since sequencing theoretically only covered coding regions, the mean editing ratios among the ~5,000 sites were globally lower, but there was nevertheless a trend in that tumor samples were more heavily edited than normal samples (Figure 3B). When we focused on editing sites in coding regions, we again observed higher editing ratios in tumors compared to normal samples (Figure 3C,D). Since we have shown that the editing sites are distinctly classified into Alu and non-Alu categories, we should separately compare the patterns in these two groups of sites. We found that for the editing sites in Alu elements and coding genes, the editing ratio was globally higher in tumors than in normal samples (Figure 4A,B).
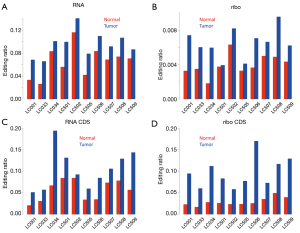
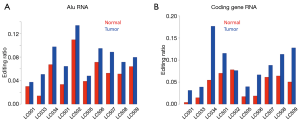
So far, we have demonstrated that editing ratios are globally elevated in tumor samples. The reason for this change, possibly related to the editing enzymes, remains to be explored.
ADARs are highly expressed and translated in tumor samples
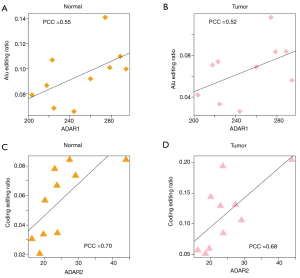
The globally elevated editing ratios in tumor samples prompted us to consider whether this was caused by regulation of the editing enzyme, ADAR. We have shown that humans have three ADARs. Previous studies have reported that ADAR3 is inactive, while ADAR1 and ADAR2 are responsible for the global editome (7). We investigated the expression level (RNA-Seq and ribo-seq) and TE of the three ADAR genes (Figure 5). In the ten patients, ADAR1 and ADAR2 levels were upregulated in tumors at both the mRNA and protein levels, whereas ADAR3 did not exhibit significant differences between tumor and normal samples (Figure 5). This observation further confirmed the higher editing efficiency of tumors. Moreover, it has been reported that the editing efficiency of Alu sites is primarily determined by ADAR1, while the editing efficiency of sites in coding regions is mainly determined by ADAR2 (7). Hence, we were curious as to whether ADAR expression and editing efficiency were correlated among the 10 patients. We tested the following correlations: ADAR1 expression versus Alu editing ratios in ten normal tissues (Figure 6A), ADAR1 expression versus Alu editing ratios in ten tumor tissues (Figure 6B), ADAR2 expression versus coding gene editing ratios in ten normal tissues (Figure 6C), and ADAR2 expression versus coding gene editing ratios in ten tumor tissues (Figure 6D). A significant positive correlation was found in all of these comparisons, suggesting that our quantification of editing ratios and ADAR expression were accurate.
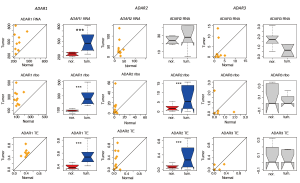
However, we still speculated as to why ADAR genes are upregulated in tumors. Since ADAR1 exhibited higher expression than ADAR2, we sought potential determinants accounting for the ADAR1 expression/translation change. Interestingly, ADAR proteins can edit mRNAs. ADAR1 pre-mRNA has several self-editing sites in its introns. Based on the GU-AG splicing rule, intronic A-to-I editing has the potential to create or abolish RNA splicing sites (Figure 7A). Gain or loss of splicing sites could lead to truncated or lengthened RNAs, and length could be a factor affecting gene expression or translation. If ADAR expression needs to be stabilized, it should avoid editing its introns. Intriguingly, for all of the intronic editing sites recorded in databases (12), 36% (699,007/1,936,298) of them would cause gain or loss of RNA splice sites, whereas for ADAR1, only 20% (10/51) of intronic sites would result in splicing gain or loss. This indicates that the editing enzyme tried to stabilize its own expression level by avoiding editing the splicing regions in its own intron. However, we still found two editing sites in the ADAR1 intron (chr1:154586049 and chr1:154584974) that had higher editing ratios in all tumor-normal pairs (Figure 7B). More strikingly, these two intronic sites caused a gain in splicing sites. That is to say, under normal conditions, these two sites were not edited so that the introns were retained. However, in tumor samples, these two sites were edited, leading to intron excision. ADAR1 mRNA was shortened in tumors. Since mRNA length was negatively correlated with TE (Figure 7C), the shortened ADAR1 RNA in tumors should lead to higher TE. This might explain the observed elevated ADAR1 levels in tumors compared to those found in normal samples.

Note that the most intuitive logic is to seek editing sites in coding regions or missense editing sites that change the mRNA-encoded amino acid sequence. However, in the case of the ADAR gene, no self-editing sites in coding regions were found. ADAR only edits its own introns. Moreover, we required the editing ratios to be higher in all ten pairs of tumor versus normal comparisons. Only two intronic self-editing sites met this stringent criterion.
Editing in oncogenes and TSG might also affect gene expression
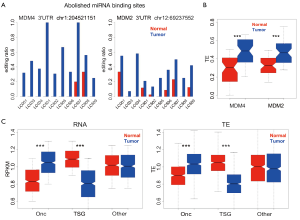
We identified that editing efficiency is globally elevated in tumor samples compared to normal samples (Figure 3). Next, we confirmed that oncogenes and TSGs also had higher editing ratios in tumor samples than in normal samples (P value =1.4e-5 and 7.5e-7 calculated by KS tests). Among oncogenes and TSGs, we searched for editing sites with editing ratios higher in all ten tumor samples. We found two such sites, chr1:204521151 and chr12:69237552 (Figure 8A). The former was in oncogene MDM4, and the latter was in oncogene MDM2. Both such editing sites were located in 3'-UTRs and abolished miRNA binding sites (Figure 8A). The binding of microRNAs represses the expression and translation of target genes, so the abolishment of microRNA binding sites might enhance gene expression and translation. Indeed, the TEs of both MDM4 and MDM2 were higher in tumors than in normal samples (Figure 8B). Next, we asked whether the observed expression or translation changes were a general pattern for oncogenes and TSGs. As anticipated, oncogenes were upregulated at the expression and translation levels in tumors, while TSGs genes were downregulated in tumors (Figure 8C). This finding might be associated with editing changes involving alterations to the expression of host genes.
Discussion
Cellular homeostasis is maintained by numerous mechanisms. Conversely, the abnormal behavior of cells, such as proliferation by tumor cells, can also be attributed to dysfunction involving multiple processes. In cancer studies, only gene expression alterations were obviously insufficient. The cis elements and trans factors such as deleterious mutations (37), RNA/protein modifications (38), codon biases (39), splicing changes (40), and microRNA targeting (41) also directly or indirectly regulate many biological processes, contributing to the maintenance or disruption of homeostasis. By comparing tumor and matched normal samples, one could always identify multiple genes that were found to be significantly upregulated or downregulated. However, the detailed molecular mechanisms underlying the observed changes in expression need to be further explored.
In this study, we found that global editing ratios were elevated in tumor samples compared to normal samples. We did not simply present this result as an observation. Instead, we associated enhanced editing efficiency with up-regulation of the editing enzyme, ADAR. Furthermore, why are ADAR genes upregulated in tumors? There are additional determinants governing this process. We found that the ratios of two self-editing sites in the ADAR1 intron that affect mRNA splicing were upregulated in tumors. Again, the gain of splicing sites resulting in abbreviated mRNAs explained the observed elevated ADAR expression and translation. This is a feed-forward loop involving ADAR (Figure 9). Higher ADAR protein levels would lead to even more extensive self-editing involving ADAR RNA introns, and such elevated splicing efficiency would further upregulate ADAR translation as well as the amount of ADAR protein subsequently generated.

Next, the upregulated ADAR proteins not only targeted their own mRNAs but also dramatically increased the global editing ratios of other target genes, including oncogenes and TSGs (Figure 9). Editing-mediated loss of miRNA-binding sites could enhance the expression of oncogenes. Meanwhile, editing-mediated splicing changes might also decrease the expression of TSGs. Both consequences would result in oncogenesis. Therefore, we propose a potential pathway that explains most nodes in the chain of oncogenesis. The only unexplained point was “the first move” at the very beginning of oncogenesis, that is, what caused the initially higher editing ratio involving the intronic editing sites in ADAR transcripts?
In summary, our study clarified the role of many biological processes and pathways in oncogenesis. We described oncogenesis from novel aspects. We stress that not only gene expression patterns should be investigated in cancer studies. The findings from our work should be appealing to the broad cancer community.
Conclusions
A-to-I RNA editing plays a crucial role in the oncogenesis of liver cancer. ADAR regulates its own expression by self-editing, and also affects the global transcriptome and translatome of cancer-related genes by editing and changing their expression patterns.
Acknowledgments
We thank colleagues who gave precious suggestions to this study. We thank the Wiley Editing Service for language editing.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-236
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-236). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet 2001;2:869-78. [Crossref] [PubMed]
- Eisenberg E, Levanon EY. A-to-I RNA editing - immune protector and transcriptome diversifier. Nature Reviews Genetics 2018;19:473-90. [Crossref] [PubMed]
- Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biology 2012;13:252. [Crossref] [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 2017;27:1115-27. [Crossref] [PubMed]
- Li Z, Weng H, Su R, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)- Methyladenosine RNA Demethylase. Cancer Cell 2017;31:127-41. [Crossref] [PubMed]
- Tan MH, Li Q, Shanmugam R, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017;550:249-54. [Crossref] [PubMed]
- Palladino MJ, Keegan LP, O'Connell MA, et al. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. Rna 2000;6:1004-18. [Crossref] [PubMed]
- Keegan LP, McGurk L, Palavicini JP, et al. Functional conservation in human and Drosophila of Metazoan ADAR2 involved in RNA editing: loss of ADAR1 in insects. Nucleic Acids Res 2011;39:7249-62. [Crossref] [PubMed]
- Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun 2011;2:319. [Crossref] [PubMed]
- Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics 2010;26:1772-6. [Crossref] [PubMed]
- Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 2014;42:D109-13. [Crossref] [PubMed]
- Li Y, Yang X, Wang N, et al. Mutation profile of over 4500 SARS-CoV-2 isolations reveals prevalent cytosine-to-uridine deamination on viral RNAs. Future Microbiol 2020;15:1343-52. [Crossref] [PubMed]
- Li Y, Yang XN, Wang N, et al. The divergence between SARS-CoV-2 and RaTG13 might be overestimated due to the extensive RNA modification. Future Virology 2020;15:341-7. [Crossref]
- Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 2014;24:365-76. [Crossref] [PubMed]
- Picardi E, Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics 2013;29:1813-4. [Crossref] [PubMed]
- Xu G, Zhang J. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A 2014;111:3769-74. [Crossref] [PubMed]
- Yu Y, Zhou H, Kong Y, et al. The Landscape of A-to-I RNA Editome Is Shaped by Both Positive and Purifying Selection. PLoS Genet 2016;12:e1006191. [Crossref] [PubMed]
- Liscovitch-Brauer N, Alon S, Porath HT, et al. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017;169:191-202 e11. [Crossref] [PubMed]
- Chu D, Wei L. The chloroplast and mitochondrial C-to-U RNA editing in Arabidopsis thaliana shows signals of adaptation. Plant Direct 2019;3:e00169. [Crossref] [PubMed]
- Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature 1999;399:75-80. [Crossref] [PubMed]
- Hsiao YE, Bahn JH, Yang Y, et al. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Research 2018;28:812-23. [Crossref] [PubMed]
- Licht K, Kapoor U, Mayrhofer E, et al. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Research 2016;44:6398-408. [Crossref] [PubMed]
- Shen S, Park JW, Lu ZX, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 2014;111:E5593-E5601. [Crossref] [PubMed]
- Chu D, Wei L. Characterizing the heat response of Arabidopsis thaliana from the perspective of codon usage bias and translational regulation. J Plant Physiol 2019;240:153012. [Crossref] [PubMed]
- Li L, Song Y, Shi X, et al. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res 2018;28:132-43. [Crossref] [PubMed]
- Wang N, Wang D. Genome-wide transcriptome and translatome analyses reveal the role of protein extension and domestication in liver cancer oncogenesis. Mol Genet Genomics 2021;296:561-9. [Crossref] [PubMed]
- Kochetov AV, Sarai A, Rogozin IB, et al. The role of alternative translation start sites in the generation of human protein diversity. Mol Genet Genomics 2005;273:491-6. [Crossref] [PubMed]
- Li Q, Li J, Yu CP, et al. Synonymous mutations that regulate translation speed might play a non-negligible role in liver cancer development. BMC Cancer 2021;21:388. [Crossref] [PubMed]
- Zhao S, Song S, Qi Q, et al. Cost-efficiency tradeoff is optimized in various cancer types revealed by genome-wide analysis. Mol Genet Genomics 2021;296:369-78. [Crossref] [PubMed]
- Beghini A, Ripamonti CB, Peterlongo P, et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 2000;9:2297-304. [Crossref] [PubMed]
- Gallo A, Vukic D, Michalik D, et al. ADAR RNA editing in human disease; more to it than meets the I. Hum Genet 2017;136:1265-78. [Crossref] [PubMed]
- Zou Q, Xiao Z, Huang R, et al. Survey of the translation shifts in hepatocellular carcinoma with ribosome profiling. Theranostics 2019;9:4141-55. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166-9. [Crossref] [PubMed]
- Chang S, Li J, Li Q, et al. Retrieving the deleterious mutations before extinction: genome-wide comparison of shared derived mutations in liver cancer and normal population. Postgrad Med J 2021; Epub ahead of print. [Crossref] [PubMed]
- Fung TS, Liu DX. Post-translational modifications of coronavirus proteins: roles and function. Future Virol 2018;13:405-30. [Crossref] [PubMed]
- Arella D, Dilucca M, Giansanti A. Codon usage bias and environmental adaptation in microbial organisms. Mol Genet Genomics 2021;296:751-62. [Crossref] [PubMed]
- Liu Q, Hu H, Wang H. Mutational bias is the driving force for shaping the synonymous codon usage pattern of alternatively spliced genes in rice (Oryza sativa L.). Mol Genet Genomics 2015;290:649-60. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]


