
Overexpression of microRNA-19a-3p promotes lymph node metastasis of esophageal squamous cell carcinoma via the RAC1/CDC42-PAK1 pathway
Introduction
Esophageal cancer (EC) is an aggressive disease that ranks eighth in incidence and sixth in tumor-related mortality worldwide (1). Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of EC and accounts for more than 90% of EC cases, especially in China (2). Regional lymph node metastasis (LNM) is a typical clinical characteristic of ESCC (3), and the incidence of regional LNM is 40–70% (3,4). ESCC is characterized by a wide lymphatic drainage region that extends from the lower neck to the upper abdomen. These features are associated with difficulties in the treatment of ESCC for the control of regional LNM. The cure rate of ESCC is relatively low, and the 5-year overall survival rate for ESCC patients is <50%, which can be partly attributed to the characteristics of regional LNM (5,6). Therefore, research efforts have focused on understanding the molecular mechanism of regional LNM and on identifying tumor-specific biomarkers as the predictor of regional LNM to guide the treatment of ESCC. However, the molecular mechanisms underlying regional LNM remain to be elucidated, and tumor-specific biomarkers for the prediction of regional LNM have not been identified to date.
MicroRNAs (miRNAs) are endogenously expressed small noncoding RNAs of 18–24 nucleotides that can modulate gene expression at the post-transcriptional level (7,8). Accumulating evidence indicates that miRNAs are involved in various biological processes in cancer, including tumor differentiation, proliferation, apoptosis, invasion, and metastasis (9-11). miRNAs have tumor-specificity and can serve as biomarkers for tumor diagnosis and prognosis (12,13). In ESCC, miRNAs are also involved in a variety of tumor-related biological processes, including proliferation, invasion, and metastasis (14-16). Although many miRNAs associated with regional LNM in ESCC have been identified (17-19), the conclusions are mostly based on in vitro experiments or small sample reports of clinical cases. There is a lack of specific studies based on the regional LNM status of ESCC, as well as studies focused on the functions and molecular mechanisms; therefore, the value of miRNAs for clinical application remains ambiguous, and there are currently no tumor-specific miRNAs that can be used as the predictor of regional LNM in ESCC.
In a previous microarray analysis of miRNAs related to LNM status in ESCC (20), we showed that the expression level of miR-19a-3p was significantly higher in cancer tissues with LNM than in those without. A verification study based on clinical cases showed that miR-19a-3p upregulation is associated significantly with LNM and may serve as a tumor-specific miRNA for the prediction of regional LNM in ESCC. However, the functions and underlying mechanisms remain to be elucidated. In this study, we expanded on the findings of our previous clinical study and performed in vitro experiments to elucidate the functions and mechanisms of miR-19a-3p in the regulation of regional LNM in ESCC. We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/tcr-21-254).
Methods
Cell culture and lentiviral transfection
The human ESCC cell lines KYSE-150 and TE-1, which presented relatively high expression of miR-19a-3p in preliminary experiments, were selected. These cell lines were purchased from Genechem (Shanghai, China) and cultured in RPMI-1640 medium (GIBCO, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, HyClone, Logan, USA) in a humidified incubator containing 5% CO2 at 37 °C. Cells in good growth state were collected for subsequent experiments.
Based on the upregulated expression of miR-19a-3p in the cell lines, which were consistent with our previous study, a miR-19a-3p inhibition sequence and a negative control sequence were used to transfect experimental cell lines. Lentiviral vectors for miR-19a-3p inhibition (LV-miR-19a-3p-inhibition) and a negative control (LVcon137) were constructed by Genechem and used to infect KYSE-150 and TE-1 cell lines. Lentiviral transfection, LV-miR-19a-3p-inhibition, or LVcon137 and KYSE-150 or TE-1 cell suspension at a density of 5×105 cells/mL were co-cultured in 6-well plates (Corning, NY, USA) for 16 h, and then the medium containing the virus supernatant was replaced with complete medium containing 10% FBS. After 72 h, green fluorescence signals were observed with a fluorescence microscope (Olympus, Tokyo, Japan). Cells in good growth state in which >70% of cells showed positive green fluorescence, which represented transfected cells, were used in subsequent experiments. Cells infected by LV-miR-19a-3p-inhibition and LVcon137 were defined as the KD group and NC group, respectively.
The efficacy of lentiviral transfection was verified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The relative expression level of miR-19a-3p was calculated using the comparative 2−∆∆Ct method and U6 was used as a reference. The primers were synthesized by Genechem and the sequences were as follows: miR-19a-3p forward primer: 5'-TGTGCAAATGTATGCAA-3' and reverse primer: 5'-GTGCAGGGTCCGAGGT-3'; U6 forward primer: 5'-CTCGCTTCGGCAGCACA-3' and reverse primer: 5'-AACGCTTCACGAATTTGCGT-3'.
Cell proliferation assay
The cell counting kit-8 (CCK-8) assay was used to evaluate the proliferation capabilities of cells and performed using the Enhanced CCK-8 kit (Beyotime, Shanghai, China) according to the manufacturer’s protocols. Briefly, 100 µL of KYSE-150 or TE-1 cell suspension for different groups were seeded in 96-well culture plates (Corning) at a density of 2×103 cells/well and three repeats per group. The plates were cultured for different times (days 1, 2, 3, 4, and 5). Before the culture was terminated, 10 µL of CCK-8 solution was added to each well and cultured for 4 h. Absorbance at 450 nm was measured with a spectrophotometer (Tecan, Mannedorf, Switzerland) and expressed as the optical density (OD).
Cell invasion assay
The Transwell invasion assay with Corning® BioCoat™ Matrigel® Invasion Chambers (Corning) was used to investigate the invasion capabilities of cells cultured in 24-well Transwell plates (Corning) following the manufacturer’s protocols. KYSE-150 or TE-1 cell suspension in serum-free medium was seeded in the upper chambers (coated in Matrigel) at a density of 105 cells/well. The lower chambers were filled with culture medium containing 30% FBS as a chemoattractant. Cells were cultured at 37 °C for 24 h, and non-invaded cells on the top of the Transwell were removed with a cotton swab. The invaded cells at the bottom of the Transwell were fixed with methanol and stained with Crystal Violet for 15 min, followed by counting with a light microscope at ×200 magnification. Nine random fields were selected to measure per plate, and all experiments were performed three times in different groups.
Cell migration assay
The Transwell migration assay and Celigo scratch assay were performed to investigate the migration capabilities of cells. The Transwell migration assay was performed as described for the invasion assay except that Matrigel was not added and cells were cultured at 37 °C for 16 h.
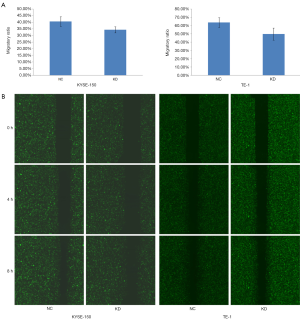
For the Celigo scratch assay, KYSE-150 or TE-1 cells were seeded in 96-well plates at a density of 5×104 cells/well with five repeats in 100 µL complete medium and cultured at 37 °C overnight. When the cultures reached 90% confluency, the cell layers were scratched with a 96-wounding replicator (VP scientific), washed three times with PBS to remove debris, and then cultured at 37 °C in complete medium containing 1% FBS. Celigo (Nexcelom) was used to scan the 96-well plates at 0, 4, and 8 h, and the migratory ratio of experimental cells was calculated. All experiments were performed five times in different groups.
Target gene assay and qRT-PCR verification
To explore the mechanisms underlying the effects of miR-19a-3p and to identify target genes, sequencing analyses were performed in KD and NC KYSE-150 cells. Total RNA was extracted from three pairs of NC and KD cells using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. To ensure that total RNA reached the requirements for the experiment, the Nano Drop ND-1000 Spectrophotometer (Thermo Fisher Scientific) was used to quantify the purity and concentration of RNA, and the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was used to assess the integrity of RNA. The sequencing analysis was performed by Genechem.
The data of sequencing analyses were combined with a literature search to identify significantly differentially expressed genes associated with tumor metastasis between KD and NC cells. Then, qRT-PCR was performed to verify these significantly differentially expressed genes. This verification was performed in the KYSE-150 and TE-1 cell lines. Relative gene expression levels were calculated using the comparative 2−∆∆Ct method, and U6 was used as a reference. All gene primers were synthesized by Invitrogen. Finally, genes showing consistent differential expression in the sequencing analysis and verification analysis in KYSE-150 and TE-1 cells were identified as the target genes for Western blot analysis.
Western blot analysis
Proteins extracted from KD and NC KYSE-150 or TE-1 cells were separated by SDS-polyacrylamide gel electrophoresis and electrotransferred onto a polyvinylidene difluoride membrane (Pall Corp, NY, USA). Then, the membrane was blocked with 5% skimmed milk for 2 h and incubated using primary antibodies against target proteins at 4 °C overnight, followed by incubation with the corresponding secondary antibodies for 2 h. Finally, the bands were visualized by electrochemiluminescence (ECL) and β-actin was used as a reference.
Statistical analysis
SPSS 20.0 software (IBM SPSS Statistics for Windows, Armonk, NY, USA) was used to perform the statistical analysis. The expression levels of genes, OD450 values, average number of invasive and migratory cells, and migratory ratio were expressed as the mean ± standard deviation (SD). The differences between two groups were compared with the independent-samples t-test. All P values were two-sided, and a value of P<0.05 was considered statistically significant.
Results
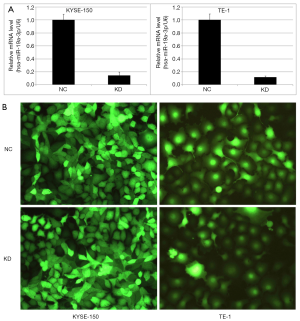
Transfection with LV-miR-19a-3p-inhibition downregulates miR-19a-3p expression
qRT-PCR analysis of cells transfected with LV-miR-19a-3p-inhibition or LVcon137 showed that the relative mRNA level of miR-19a-3p was lower in KD cells than in NC cells in both KYSE-150 and TE-1 cells (Figure 1). The P values were 1.32E-04 in KYSE-150 cells and 8.34E-05 in TE-1 cells. These results indicated that LV-miR-19a-3p-inhibition or LVcon137 transfections were effective and guaranteed the reliability of subsequent experiments.
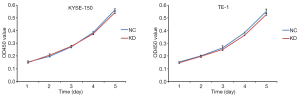
miR-19a-3p does not affect the proliferation of ESCC cells
A CCK-8 assay was used to evaluate the effect of miR-19a-3p on the proliferation of KYSE-150 or TE-1 cells. The OD450 values obtained at five time points did not differ between the KD group and NC group in KYSE-150 and TE-1 cells (Figure 2). The P values on days 1–5 were 0.7336, 0.3516, 0.8074, 0.4069, and 0.1533 in KYSE-150 cells and 0.6252, 0.7299, 0.4421, 0.1536, and 0.1296 in TE-1 cells. These results indicated that miR-19a-3p had no effect on ESCC cell proliferation.
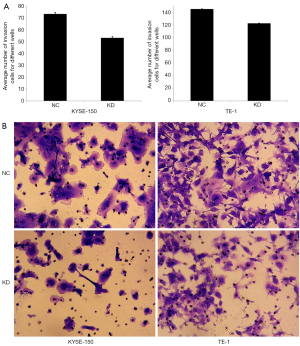
miR-19a-3p regulates the invasion of ESCC cells
The Transwell invasion assay was used to evaluate the effect of miR-19a-3p on the invasion of KYSE-150 and TE-1 cells. The results showed that the average number of invasive cells was higher in the NC group than in the KD group in KYSE-150 (P=3.21E-05) and TE-1 cells (P=1.52E-06) (Figure 3). These results indicate that LV-miR-19a-3p-inhibition decreased the invasive capabilities of ESCC cells, suggesting that overexpression of miR-19a-3p promotes the invasion of ESCC cells.
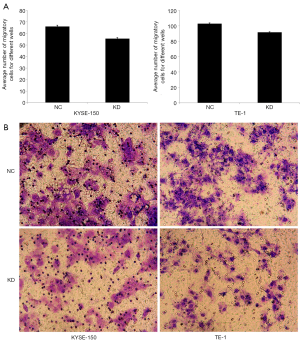
miR-19a-3p regulates the migration of ESCC cells
The Transwell migration assay and Celigo scratch assay were performed to evaluate the effect of miR-19a-3p on the migration of KYSE-150 and TE-1 cells. The results of the Transwell migration assay showed that the average number of migratory cells was higher in the NC group than in the KD group in KYSE-150 (P=2.48E-04) and TE-1 cells (P=7.52E-05) (Figure 4). The results of the Celigo scratch assay showed that the migratory ratio was higher in the NC group than in the KD group in KYSE-150 (P=0.0100) and TE-1 cells (P=0.0097) (Figure 5). These results indicate that LV-miR-19a-3p-inhibition reduced the migratory capabilities of ESCC cells, suggesting that overexpression of miR -19a-3p promotes the migration of ESCC cells.
Identification of target genes of miR-19a-3p and qRT-PCR verification
The results of sequencing analysis identified 140 significantly differentially expressed genes in KYSE-150 cells transfected with LV-miR-19a-3p-inhibition compared with LVcon137 according to a standard of Padj<0.05. Among them, 30 genes were upregulated and 110 genes were downregulated in KYSE-150 cells infected with LV-miR-19a-3p-inhibition compared with LVcon137. Combined with literature reports, ten genes associated with tumor metastasis were screened for qRT-PCR verification, including two upregulated genes and eight downregulated genes. In the verification, each gene was tested three times in KYSE-150 and TE-1 cells, and three genes showing consistent differential expression in the sequencing and verification analyses in KYSE-150 and TE-1 cells were identified as the target genes of miR-19a-3p for Western blot analysis. The three target genes included cell division cycle 42 (CDC42), Rac family small GTPase 1 (RAC1), and p21 activated kinase 1 (PAK1). The three genes were downregulated in the KD group compared with the NC group in KYSE-150 and TE-1 cells (Table 1). These results indicate that miR-19a-3p regulates the expression of CDC42, RAC1, and PAK1. Western blot analysis was performed to confirm this mechanism.
Table 1
| Gene name | KYSE-150 | TE-1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KD (mean ± SD) | NC (mean ± SD) | Expression tendency | P value | Consistency with sequencing data | KD (mean ± SD) | NC (mean ± SD) | Expression tendency | P value | ||
| SP1 | 3.4543±0.7505 | 2.5049±0.5189 | Up | 0.1458 | Inconsistent | 0.3954±0.1349 | 1.3962±0.1422 | Down | 0.0009 | |
| CDH1 | 1.0490±0.1152 | 0.6027±0.1408 | Up | 0.0132 | Consistent | 1.3047±0.3826 | 0.9626±0.0996 | Up | 0.2084 | |
| STAT1 | 2.2776±0.4892 | 1.9113±0.5323 | Up | 0.4298 | Consistent | 1.1725±0.2287 | 0.6373±0.1543 | Up | 0.0283 | |
| GNA13 | 1.3604±0.4121 | 1.7661±0.4582 | Down | 0.3178 | Consistent | 1.8897±0.2478 | 1.9801±0.3525 | Down | 0.7348 | |
| PIK3C2A | 2.0363±0.5501 | 3.4991±0.4295 | Down | 0.0222 | Consistent | 5.3436±0.2610 | 4.5300±0.5236 | Up | 0.0737 | |
| PRKCA | 1.2215±0.2131 | 2.9382±0.3749 | Down | 0.0023 | Consistent | 2.1196±0.7710 | 2.8624±0.5679 | Down | 0.2502 | |
| EGR1 | 0.5606±0.3436 | 1.0297±0.3131 | Down | 0.1554 | Consistent | 0.6341±0.1025 | 1.0320±0.3310 | Down | 0.1176 | |
| CDC42 | 0.5326±0.1228 | 2.0274±0.1561 | Down | 0.0002 | Consistent | 0.2765±0.1302 | 1.4215±0.2384 | Down | 0.0019 | |
| RAC1 | 0.6651±0.1512 | 2.8659±0.3149 | Down | 0.0004 | Consistent | 0.3951±0.0836 | 2.6448±0.4216 | Down | 0.0008 | |
| PAK1 | 1.9465±0.3003 | 5.0851±0.3942 | Down | 0.0004 | Consistent | 1.0050±0.1704 | 3.3356±0.3301 | Down | 0.0004 | |
KD: cell group was transfected by lentiviral vectors of miR-19a-3p inhibition (LV-miR-19a-3p-inhibition). NC: cell group was transfected by lentiviral vectors of negative control (LVcon137). SP1, Sp1 transcription factor; CDH1, cadherin 1; STAT1, signal transducer and activator of transcription 1; GNA13, G protein subunit alpha 13; PIK3C2A, phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha; PRKCA, protein kinase C alpha; EGR1, early growth response 1; CDC42, cell division cycle 42; RAC1, Rac family small GTPase 1; PAK1, p21 (RAC1) activated kinase 1. SD, standard deviation.
The miR-19a-3p promotes CDC42, RAC1, and PAK1 expression
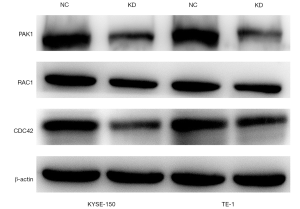
Western blot analysis showed that CDC42, RAC1, and PAK1 were downregulated in KYSE-150 and TE-1 cells transfected with LV-miR-19a-3p-inhibition, whereas they were upregulated in KYSE-150 and TE-1 cell lines transfected with LVcon137 (Figure 6). This implied that overexpression of miR-19a-3p induces the expression of CDC42, RAC1, and PAK1 to mediate its functions.
Discussion
Regional LNM is one of the most common reasons leading to treatment failure in ESCC (3,4), which results in depressing survival of ESCC patients (5,6). The lymphatic drainage region of ESCC is extensive, and a tendency toward extensive regional LNM is a typical clinical feature of ESCC (3,4). These features make it difficult to predict and control LNM in ESCC. However, in clinical practice, regional LNM differs considerably among patients. The ability to predict these differences and take effective measures to treat LNM in patients with ESCC would represent a significant advance in the treatment of this disease. However, despite extensive research on regional LNM of ESCC in the last decades (21), tumor-specific markers capable of predicting LNM and guiding the treatment of ESCC patients remain unidentified.
The miRNAs have been reported to be involved in the invasion and metastasis of ESCC, and their tumor-specificity suggests that they could serve as biomarkers for predicting invasion and metastasis of ESCC (14-16). However, few studies based on regional LNM status were performed to predict LNM and guide the treatment of ESCC. Studies show that miR-19a-3p is involved in invasion and metastasis in hepatocellular carcinoma, gastric cancer, and ovarian cancer (22-24), but its predictive value for regional LNM in ESCC has not been reported. In previous work from our group, we showed that miR-19a-3p predicts the LNM of ESCC and might act as a biomarker for LNM in ESCC (20), but the underlying mechanisms were not elucidated. In this study, we examined the functions and mechanisms of miR-19a-3p in vitro. Based on our previous study showing that miR-19a-3p is significantly upregulated in ESCC, lentiviral vectors for miR-19a-3p inhibition and a negative control were constructed to transfect experimental cell lines, and qRT-PCR was used to verify the transfection efficacy. The results showed that transfection with LV-miR-19a-3p-inhibition downregulated the expression level of miR-19a-3p.
In our previous clinical study, miR-19a-3p was only associated with LNM of ESCC (20). In this functional study in vitro, the results of functional experiments showed that miR-19a-3p had no influence on the proliferation of ESCC cells; however, overexpression of miR-19a-3p promoted invasion and migration of ESCC cells. These results confirmed that miR-19a-3p has the potential to promote the LNM of ESCC, which is consistent with our previous clinical study.
After the functional experiments, we examined the mechanisms underlying the effect of miR-19a-3p on promoting the invasion and migration of ESCC cells. The mechanistic studies consisted of sequencing analysis and IPA based on KD and NC of KYSE-150 cells. The sequencing analysis and IPA identified significantly differentially expressed genes regulated by miR-19a-3p and related pathways associated with tumor metastasis, which guaranteed the smooth implementation and the reliability of subsequent study of the mechanism. Ten differentially expressed genes from five pathways associated with tumor metastasis were selected for qRT-PCR verification in KYSE-150 and TE-1 cells. Three genes including CDC42, RAC1, and PAK1, which were downregulated in LV-miR-19a-3p-inhibition transfected cells, were confirmed to be differentially expressed in the two cell lines, and the expression tendency was consistent with the sequencing analysis in KYSE-150 cells. The three genes were identified as the target genes of miR-19a-3p. Western blot analysis confirmed that the three target genes were downregulated in KYSE-150 and TE-1 cells infected with LV-miR-19a-3p-inhibition. These results indicate that overexpression of miR-19a-3p may facilitate the invasion and migration of ESCC cells by regulating the expression of CDC42, RAC1, and PAK1.
CDC42 is a small GTPase of the Rho-subfamily, which is involved in many signaling pathways that regulate diverse biological functions, including cell migration, endocytosis, and cell cycle progression (25,26). RAC1 is a cell membrane-related small GTPase; in its active form, it binds to various effector proteins to regulate cellular processes such as secretory processes, neuron adhesion, epithelial cell polarization, phagocytosis of apoptotic cells, differentiation, and migration, and growth-factor induced formation of membrane ruffles (25,26). In many cancers, upstream molecules are often via activating RAC1 and CDC42 to regulate downstream effector proteins to exert their biological functions, including cell migration (27-29). For example, Xu et al. reported that POTEE facilitated colorectal cancer cell migration by activating the RAC1/CDC42 axis (27). Chang et al. reported that GIT1 facilitated lung cancer cell metastasis by regulating RAC1/CDC42 activity (28). Wang et al. reported that overexpression of TEM8 facilitated progression of ovarian cancer via the RAC1/CDC42 pathway (29).
PAK1 is a serine/threonine protein kinase that has a crucial role in various signaling pathways and often acts as downstream effector of the small GTPases CDC42 and RAC1 (30). CDC42 and RAC1 bind to PAK1, causing a conformational transform and the subsequent autophosphorylation of several serine and/or threonine residues, which activates PAK1 to exert its biological functions including cell migration (30). PAK1 is involved in almost every stage of the cell migration process including facilitating angiogenesis via VEGF-mediated Raf-1 and MEK1 activation during angiogenesis (31), promoting tumor epithelial-mesenchymal transformation (EMT) by upregulating the expression of matrix metalloproteinase and activating various transcription factors (32,33), and regulating the cytoskeleton remodeling process to promote cell migration (34). Liu et al. reported that GEFT promotes rhabdomyosarcoma cell migration via the RAC1/CDC42-PAK1 pathways to regulate EMT (35). In this study, transfection with LV-miR-19a-3p-inhibition downregulated the expression of RAC1, CDC42, and PAK1, suggesting that the RAC1/CDC42-PAK1 signaling pathway is involved in the regulation of miR-19a-3p function. The results suggest that miR-19a-3p regulates ESCC cell migration via the RAC1/CDC42-PAK1 signaling pathway. Taken together with previous results showing that miR-19a-3p promotes regional LNM of ESCC, the present findings suggest that miR-19a-3p promotes regional LNM of ESCC potentially via the RAC1/CDC42-PAK1 signaling pathway, but further studies are necessary to elucidate the detailed underlying mechanisms in the future.
Conclusions
In summary, the results of in vitro experiments showed that overexpression of miR-19a-3p promotes invasion and migration of ESCC cells via the RAC1/CDC42-PAK1 signaling pathway, suggesting that miR-19a-3p promotes regional LNM of ESCC potentially via this mechanism. Further studies are necessary to explore the detailed underlying mechanisms.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-254
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-254
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-254). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Wang Y, Zhu L, Xia W, et al. Anatomy of lymphatic drainage of the esophagus and lymph node metastasis of thoracic esophageal cancer. Cancer Manag Res 2018;10:6295-303. [Crossref] [PubMed]
- Tachimori Y. Pattern of lymph node metastases of squamous cell esophageal cancer based on the anatomical lymphatic drainage system: efficacy of lymph node dissection according to tumor location. J Thorac Dis 2017;9:S724-30. [Crossref] [PubMed]
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Rustgi A, El-Serag HB. Esophageal carcinoma. New Engl J Med 2015;372:1472-3. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006;20:515-24. [Crossref] [PubMed]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006;94:776-80. [Crossref] [PubMed]
- Lopez-Camarillo C, Marchat LA, Arechaga-Ocampo E, et al. MetastamiRs: non-coding miRNAs driving cancer invasion and metastasis. Int J Mol Sci 2012;13:1347-79. [Crossref] [PubMed]
- Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci 2015;22:9. [Crossref] [PubMed]
- Jay C, Nemunaitis J, Chen P, et al. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol 2007;26:293-300. [Crossref] [PubMed]
- He B, Zhao Z, Cai Q, et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci 2020;16:2628-47. [Crossref] [PubMed]
- Feng Q, Zhang H, Yao D, et al. Emerging Role of Non-Coding RNAs in Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2019;21:258. [Crossref] [PubMed]
- Mei LL, Qiu YT, Zhang B, et al. MicroRNAs in esophageal squamous cell carcinoma: Potential biomarkers and therapeutic targets. Cancer Biomark 2017;19:1-9. [Crossref] [PubMed]
- Harada K, Baba Y, Ishimoto T, et al. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol 2016;51:520-30. [Crossref] [PubMed]
- Liu T, Du LT, Wang YS, et al. Development of a Novel Serum Exosomal MicroRNA Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Esophageal squamous cell carcinoma. Front Oncol 2020;10:573501. [Crossref] [PubMed]
- Liu ZH, Chen LD, He YB, et al. Study of expression levels and clinical significance of miR-503 and miR-375 in patients with esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci 2019;23:3799-805. [PubMed]
- Chen L, Wang Y, Wang W, et al. Hsa-miR-6743-5p Expression Varies with Lymph Node Metastasis in Esophageal Cancer. Clin Lab 2018;64:1249-57. [Crossref] [PubMed]
- Xu Y, Sun DS, Zhang XD, et al. Microarray analysis of miRNA based on the regional lymph node metastasis status of esophageal squamous cell carcinoma. Transl Cancer Res 2021;10:273-87. [Crossref]
- Li J, Qi Z, Hu YP, et al. Possible biomarkers for predicting lymph node metastasis of esophageal squamous cell carcinoma: a review. J Int Med Res 2019;47:544-56. [Crossref] [PubMed]
- Wang Z, Shi Z, Zhang L, et al. Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in human hepatocellular carcinoma. Pathol Res Pract 2019;215:499-505. [Crossref] [PubMed]
- Li X, Yan X, Wang F, et al. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J Cell Mol Med 2019;23:2920-32. [Crossref] [PubMed]
- Bai R, Cui Z, Ma Y, et al. The NF-κB-modulated miR-19a-3p enhances malignancy of human ovarian cancer cells through inhibition of IGFBP-3 expression. Mol Carcinog 2019;58:2254-65. [Crossref] [PubMed]
- Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016;7:207-21. [Crossref] [PubMed]
- Svensmark JH, Brakebusch C. Rho GTPases in cancer: friend or foe? Oncogene 2019;38:7447-56. [Crossref] [PubMed]
- Xu Q, Chen J, Peng M, et al. POTEE promotes colorectal carcinoma progression via activating the Rac1/Cdc42 pathway. Exp Cell Res 2020;390:111933. [Crossref] [PubMed]
- Chang JS, Su CY, Yu WH, et al. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget 2015;6:36278-91. [Crossref] [PubMed]
- Wang CX, Xiong HF, Wang S, et al. Overexpression of TEM8 promotes ovarian cancer__progression via Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways. Am J Transl Res 2020;12:3557-76. [PubMed]
- Yao D, Li C, Rajoka MSR, et al. P21-Activated Kinase 1: Emerging biological functions and potential therapeutic targets in Cancer. Theranostics 2020;10:9741-66. [Crossref] [PubMed]
- Alavi A, Hood JD, Frausto R, et al. Role of raf in vascular protection from distinct apoptotic stimuli. Science 2003;301:94-6. [Crossref] [PubMed]
- Chow HY, Dong B, Valencia CA, et al. Group i paks are essential for epithelial- mesenchymal transition in an apc-driven model of colorectal cancer. Nat Commun 2018;9:3473. [Crossref] [PubMed]
- Gonzalez-Villasana V, Fuentes-Mattei E, Ivan C, et al. Rac1/pak1/p38/mmp-2 axis regulates angiogenesis in ovarian cancer. Clin Cancer Res 2015;21:2127-37. [Crossref] [PubMed]
- Sells MA, Knaus UG, Bagrodia S, et al. Human p21-activated kinase (pak1) regulates actin organization in mammalian cells. Curr Biol 1997;7:202-10. [Crossref] [PubMed]
- Liu C, Zhang L, Cui W, et al. Epigenetically upregulated GEFT-derived invasion and metastasis of rhabdomyosarcoma via epithelial mesenchymal transition promoted by the Rac1/Cdc42-PAK signalling pathway. EBioMedicine 2019;50:122-34. [Crossref] [PubMed]


