
Analyzing microsatellite instability and gene mutation in circulating cell-free DNA to monitor colorectal cancer progression
Introduction
Colorectal cancer (CRC) is the fourth most common incident malignancy and the second leading cause of cancer deaths worldwide, accounting for 1.8 million new cases and 896,000 deaths in 2017 (1). According to the molecular genetic, pathological, and clinical characteristics, CRC has been defined into four consensus molecular subtypes (CMS): CMS1, CMS2, CMS3, and CMS4 (2). About 15% of CRCs belong to CMS1 which are characterized by high-level microsatellite instability (MSI-H) as a consequence of either a germline mutation in one of the mismatch repair (MMR) genes (Lynch syndrome, 3%) or a somatic mutation in the same pathway (sporadic MSI-H, 12%) (3). The remaining 85% of CRCs are classified as microsatellite stability (MSS) or low-level microsatellite instability (MSI-L) phenotype, developing from chromosomal changes (4). CRCs with deficient DNA MMR exhibit unique clinical, pathological, and molecular characteristics compared to those with proficient DNA MMR (5). It is reported that MSI CRCs are more likely to be proximal, medullary, mucinous, poorly differentiated, surrounded by increased tumor-infiltrating lymphocytes (6) with favorable prognosis (7) and sensitive to immune checkpoint blockade with antibodies to programmed death receptor-1 (PD-1) (8,9). MSI status is a predictor of the efficiency from adjuvant chemotherapy for stage II CRC and fluorouracil therapy alone is not recommended for patients with stage II CRCs of MSI-H phenotype (10,11). Thus, stratification by MSI status may facilitate the individualized therapy for CRC patients.
Traditionally, both polymerase chain reaction (PCR) to infer MSI with a set of mononucleotide or dinucleotide markers and immunohistochemistry to evaluate the expression of the main MMR proteins are fundamental methods for analyzing MSI status (12). Recent studies showed that next-generation sequencing (NGS) was an alternative and more practical method to determine MSI status than MSI-PCR (13-15) because it does not require matched normal tissue samples (16).
Advances in targeted therapy and chemotherapy have significantly improved the CRC patients’ overall survival (17,18). The presence of the specific mutation, such as KRAS, BRAF, and HER-2 mutation, may confer resistance to targeted therapy and affect the prognosis, genetic testing is necessary for guiding the CRC treatment. However, with the progression of cancer, heterogeneity within the primary tumor or between primary and metastasis affects the response to therapy. Repeated genetic testing is required for targeted therapy, tumor biopsy is limited due to potentially harmful procedures. Liquid biopsy refers to a method of testing for biomarkers of cancer or other diseases by analyzing circulating cell-free DNA (cfDNA), exosomes, and circulating tumor cells (CTCs) in peripheral blood. As a breakthrough technology, liquid biopsy can noninvasively detect various types of DNA alterations and predict drug resistance, in addition to monitoring tumor molecular evolution at the different stages during therapy (19,20). Previous studies showed that the sensitivity of tumor mutation detection by cfDNA in metastatic CRC patients was nearly 90% (21-23). However, the feasibility of mutation testing in cfDNA from the resectable CRC patients remains unknown. In the present study, we have compared the MSI status and mutation profiles of surgically resected CRC tissues with those of matched preoperative plasma samples to investigate the clinical value of liquid biopsy in detecting MSI status and reflecting molecular characteristics of CRC. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) checklist (available at https://dx.doi.org/10.21037/tcr-20-2762).
Methods
Patients
Plasma samples and matched primary CRC tissues were obtained from 6 patients who underwent surgery for CRC at Nanfang Hospital (Guangzhou, China) between October and November 2016. Patients were assessed pathologically after surgery using the TNM system (24) and histopathologic examination revealed all tumors to be colorectal adenocarcinomas. Tumor volume was calculated as (length × width2)/2 (25). Clinical data including age, sex, level of carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9), and pathology parameters including tumor size, grade, and stage were collected for analyses. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Nanfang Hospital (Guangzhou, China) (NFEC-2017-193). All patients signed the informed consent.
Sample collection and DNA extraction.
Peripheral blood samples were collected the day before surgery and centrifuged at 1,600 ×g for 10 min within 2 h after collection to separate plasma. Cell-free DNA was extracted from 4 to 5 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) following the manufacturer’s operational manual. Tissue samples were collected from the primary site during surgery. DNA was extracted from tumor tissue using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Immunohistochemistry staining (IHC) analysis of MMR proteins
IHC staining was performed using formalin-fixed, paraffin-embedded (FFPE) CRC tissue sections as described above. After dewaxing, antigen retrieval, and 3% hydrogen peroxide blocking nonspecific signals, the slides were incubated by primary antibodies overnight at 4 °C, then followed by the secondary antibody for 30 mins and the third antibody for 20 mins at room temperature, and DAB (3,3-diaminobenzidine) substrate staining and hematoxylin counterstaining, with the interval of three times rinsing of 0.01 mol/L phosphate-buffered saline (PBS). Finally, the slides were dehydrated and mounted. The primary antibodies included MLH1, MSH2, MSH6, and PMS2 (brand and catalogue number).
Any nuclear staining of cancer cells was considered positive and the case was defined as protein proficient. Tumors displaying proficient MMR (pMMR) proteins were considered to be microsatellite stable (MSS) or MSI-low (MSI-L). The negative nucleus staining was defined as protein deficient. The case which lost any of the four proteins was defined as deficient MMR (dMMR) and expected to be MSI-H (26). All slides were analyzed independently by two experienced pathologists who were blinded to patient clinical characteristics. Any discordant cases were reviewed together and a consensus result was reached.
MSI analysis by NGS
DNA extracted from both peripheral blood specimens and FFPE blocks were used for MSI analysis. MSI analysis was performed by NGS with a panel of 5 quasi-monomorphic mononucleotide repeats (BAT-25, BAT-26, NR21, NR24, and NR27). According to the percentage of loci with MSI, CRCs can be classified into MSI-high (MSI-H), MSI-low (MSI-L), and microsatellite stability (MSS). To be specific, MSI-H was defined if two or more markers showed instability, and MSI-L was defined if only one marker was unstable. If all 5 microsatellites showed stability, the case was defined as MSS (27).
Sequencing analysis
Tissue DNA and cfDNA samples were subjected to analysis with NGS panels for mutation detection. Genomic DNA from tumors and blood samples were fragmented and hybridized to commercially available capture arrays for enrichment. In the discovery stage, the exome capture procedure was performed with Agilent’s SureSelect Human All Exon Kit protocol (Agilent Technologies). For library preparation, tumor DNA (10 ng) and cfDNA (maximum of 3,000 copies) were subjected to multiplex PCR amplification with the use of an Ion AmpliSeq Library Kit 2.0 and Ion AmpliSeqTM Cancer Hotspot Panel v2, the latter of which consists of 207 pairs of primers that target 50 most commonly reported oncogenes and tumor suppressor genes, such as KRAS, BRAF, EGFR, and TP53. Libraries were pooled in equimolar amounts and sequenced simultaneously on the same flow cell in an Illumina Miseq sequencer (Illumina, Hayward, CA, USA). The output data was uploaded for data quality control (QC), sequence alignment, and variant calling with a vendor-supplied bioinformatics pipeline.
Statistical analysis
Statistical analysis was carried out using the two-tailed Student’s t-test (Prism software; GraphPad Software, San Diego, CA, USA). Two-sided P values less than 0.05 were considered statistically significant.
Results
Clinical characteristics
Six patients were involved in the study and their clinical characteristics were shown in Table 1. The study consisted of 2 women and 4 men with a median age at the biopsy of 59 years (range, 42–75). There were 1 Stage I tumor, 4 Stage II tumors, and 1 Stage III tumor. Plasma and MPTT samples were obtained from the above 6 patients.
Table 1
| Case | Age, years | Sex | TNM | Stage | Differentiation grade | Tumor location | Tumor volume, cm3 | Survival, years | CEA, ng/mL | CA19-9, U/mL |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 49 | M | pT2N0M0 | I | Poor | Left | 4.05 | >2 | 2.18 | 18.9 |
| 2 | 57 | M | pT4bN2aM0 | IIIC | Moderate | Rectum | 44 | >2 | 9.96 | 12.34 |
| 3 | 66 | F | pT4aN0M0 | IIB | Poor | Left | 6 | >2 | 4.75 | 8.24 |
| 4 | 75 | F | pT4aN0M0 | IIB | Poor | Right | 50.625 | >2 | NA | NA |
| 5 | 61 | M | pT4aN0M0 | IIB | Moderate | Left | 22.5 | >2 | 1.65 | 21.79 |
| 6 | 42 | M | pT3N0M0 | IIA | Moderate | Left | 48 | >2 | 0.99 | 15.89 |
Right colon is defined as a proximal colon region from the cecum and ascending colon to the right transverse colon. Left colon is defined as a distal colon region from the left transverse colon to the descending and sigmoid colon, not including the rectum. CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; NA, not available.
Concordance of cfDNA and MPTT DNA MSI status
Four in 6 patients had plasma and MPTT available for MSI status assessment by multiple methods, and these patients’ MSI status assessment has relied on NGS with MMR-IHC as reference. The other two patients didn’t have MPTT samples for MSI status detection by MSI-NGS/MMR-NGS, so their MSI status assessment was based on MMR-IHC. Comprehensive analysis of NGS and IHC results showed that the overall incidences of MSI-H in plasma and MPTT samples from these patients were 1/6 and 2/6, respectively (Table 2). Four patients were determined as microsatellite stable (MSS) in both plasma and MPTT.
Table 2
| Case | IHC | Tumor MMR | Plasma MMR | Tumor MSI | Plasma MSI |
|---|---|---|---|---|---|
| 1 | MSH2 (−), MSH6 (+), PMS2 (+), MLH1 (+) | + | − | 5/5 | 0/5 |
| 2 | MSH2 (+), MSH6 (+), PMS2 (+), MLH1 (+) | − | − | 0/5 | 0/5 |
| 3 | MSH2 (−), MSH6 (−), PMS2 (+), MLH1 (+) | − | − | 0/5 | 0/5 |
| 4 | MSH2 (+), MSH6 (+), PMS2 (−), MLH1 (−) | * | − | * | 4/5 |
| 5 | MSH2 (+), MSH6 (+), PMS2 (+), MLH1 (+) | * | − | * | 0/5 |
| 6 | MSH2 (+), MSH6 (+), PMS2 (+), MLH1 (+) | − | − | 0/5 | 0/5 |
*, indicate that the corresponding samples were unqualified. “+” “−” in IHC indicates the presence/absence of certain MMR protein, respectively. In NGS analysis of MMR, “+” indicates mutation and “−” indicates no mutation.
One patient, a 49-year-old man, had a definitive loss of MSH2 on IHC, consistent with MSI-NGS and MMR-NGS analysis of the tumor tissue sample. However, his cfDNA was defined as MSS. One was identified as MSI-H for the absence of expression of PMS2 and MLH1 in primary tumor tissue, verified in plasma by MSI-NGS analysis, but demonstrated no MMR gene mutation in plasma. To conclude, in the above 6 patients, MSI-NGS analysis of cfDNA accurately identified 1/2 of tissue MSI-H and 4/4 of tissue MSS for an overall accuracy of 5/6.
Mutation profile of plasma and MPTT by NGS analysis
In the study, tissue DNA extracted from 4 patients was successfully sequenced. 23 mutant genes were detected in these tumor tissue samples. Among them, 9 genes were mutated in at least 1 tissue sample while 14 genes were mutated in only 1 tissue sample. Frequent mutant genes included FBXW7 (4/4), LRP1B (3/4), KRAS (3/4). Circulating cfDNA extracted from 6 patients was successfully sequenced. Only a small number of non-synonymous somatic mutations were identified in cfDNA. APC and KRAS mutation were found to share between plasma and MMPT sample from one 57-year-old patient. SYNE1, RNF43, PIK3CA, EGFR, and BRAF mutations were detected in a plasma sample from one 75-year-old woman whose MMPT sample was unqualified for NGS analysis. The remaining four cases showed no mutation in cfDNA. Detailed data for each gene are shown in Figure 1, Table 3.

Table 3
| Case | Gene | AA mutation | Variant allele frequency% | |
|---|---|---|---|---|
| Tumor | Plasma | |||
| 1 | ERBB2 | p.R648Q | 21.58 | 0 |
| INHBA | p.R377H | 20.41 | 0 | |
| FBXW7 | p.R144X | 19.55 | 0 | |
| MSH2 | p.K509X | 19.35 | 0 | |
| CREBBP | p.R2151W | 17.71 | 0 | |
| LRP1B | p.G2385R | 1.47 | 0 | |
| NFE2L2 | p.G15A | 0.98 | 0 | |
| CTNNB1 | p.T40I | 0.77 | 0 | |
| CTNNB1 | p.T257I | 0.74 | 0 | |
| ROS1 | p.N1901K | 0.67 | 0 | |
| PIK3CA | p.H1065L | 0.66 | 0 | |
| MLL3 | p.D2092Y | 0.60 | 0 | |
| 2 | KRAS | p.G12D | 38.07 | 2.33 |
| APC | p.E1309X | 24.04 | 2.55 | |
| FLT1 | p.R314K | 0.98 | 0 | |
| AXIN2 | p.A734V | 0.96 | 0 | |
| FBXW7 | p.S516F | 0.85 | 0 | |
| LRP1B | p.G1755X | 0.74 | 0 | |
| SYNE1 | p.L4970F | 0.71 | 0 | |
| NFE2L2 | p.D13Y | 0.68 | 0 | |
| ROS1 | p.N1901K | 0.57 | 0 | |
| 3 | KRAS | p.G12D | 1.62 | 0 |
| PTEN | p.I101N | 0.87 | 0 | |
| FGFR4 | p.R434Q | 0.84 | 0 | |
| RNF43 | p.R132X | 0.79 | 0 | |
| FBXW7 | p.R144Q | 0.68 | 0 | |
| MLL3 | p.D2092Y | 0.65 | 0 | |
| ERBB4 | p.P700S | 0.64 | 0 | |
| LRP1B | p.S2421S | 0.62 | 0 | |
| LRP1B | p.R3922I | 0.58 | 0 | |
| EGFR | p.M766fs | 0.31 | 0 | |
| 4 | SYNE1 | p.A4961V | * | 1.53 |
| RNF43 | p.R337X | * | 1.05 | |
| PIK3CA | p.H1047R | * | 0.91 | |
| EGFR | p.A722V | * | 0.59 | |
| BRAF | p.V600E | * | 0.45 | |
| 5 | – | – | * | – |
| 6 | KRAS | p.G12D | 28.23 | 0 |
| JAK2 | p.Y114C | 0.57 | 0 | |
| BRAF | p.W531L | 0.56 | 0 | |
| SYNE1 | p.L4753Q | 0.54 | 0 | |
| FBXW7 | p.Q156H | 0.48 | 0 | |
| PIK3CA | p.Q137L | 0.47 | 0 | |
| APC | p.A697S | 0.40 | 0 | |
*, indicate that the corresponding samples were unqualified. Non-detected mutations are indicated with a dot. AA mutation: amino acid mutation.
Association of clinical characteristics with cfDNA mutation incidence
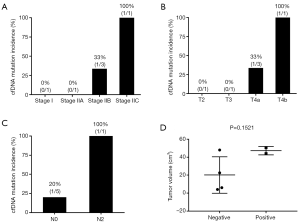
In the study, both plasma and MPTT samples defined as MSI-H consistently were obtained from a 75-year-old female patient at stage IIb (pT4a/N0/M0) and she was found to be cfDNA mutation-positive at the same time. The patient with inconsistent results of MSI status was classified as stage I (pT2/N0/M0). Another cfDNA mutation-positive case was classified as stage IIIc (pT4b/N2a/M0). Moreover, tumor volume revealed no statistically significant association with cfDNA mutation status in plasma (cfDNA mutation positive 47.31±3.313 vs. cfDNA mutation negative 20.14±10.17, P>0.05) (Figure 2). As for other markers in plasma such as CEA and CA19-9, in our study, only 1 in 6 patients’ CEA level was higher than the threshold of 5 µg/L recommended by the American Society of Clinical Oncology (28), and this patient was also found cfDNA mutation-positive.
Discussion
Comparative analyses about the sensitivity and accuracy between plasma and MPTT samples detection had done in 6 CRC patients. The study showed that MSI-NGS detection in cfDNA accurately identified 1/2 of tissue MSI-H and 4/4 of tissue MSS for an overall accuracy of 5/6, suggesting the significant clinical utility of testing MSI status in cfDNA from CRC patients. Consistently, a large cfDNA-tissue MSI concordance cohort study demonstrated that cfDNA MSI testing accurately detected 87% (71/82) of tissue MSI-H and 99.5% of tissue MSS (863/867) (29). Also, the Chinese Society of Clinical Oncology (CSCO) diagnosis and treatment guidelines for CRC have regarded MSI-NGS testing of cfDNA as an alternative to determine the MSI status of patients lacking tissue (30). These studies highlighted the potential of MSI-NGS testing of cfDNA as a noninvasive tool for monitoring the MSI status of CRC, especially the advanced CRC.
Some studies have demonstrated the clinical validity of cfDNA assays from patients with certain types of metastatic solid cancer (31-34). However, few studies have estimated the mutation status of cfDNA isolated from plasma of patients with CRC at the resectable stage. In the present study, gene mutations were detected in 4/5 of tissue samples and 2/6 of plasma samples. Moreover, the number and frequency of mutant genes detected in plasma were less than those of MPTT samples. The relationship of the cfDNA MSI and gene mutation detection with the clinical characteristics was investigated in this study. The results indicated that the sensitivity of MSI status and gene mutation detection in plasma might be positively correlated with tumor stage rather than tumor volume. The stage-dependent sensitivity of plasma markers for CRC suggested that markers releasing from the primary tumor into plasma may rely on vascular invasion (35). Therefore, the feasibility of mutation detection with cfDNA extracted from patients with resectable CRC was limited. However, it requires further validation because the sample size is too small to be conclusive.
What could explain the inconsistent results in plasma and MPTT samples? First of all, the technique may be insufficiently sensitive because the characterization of MSI and mutation profile from the tumor-specific DNA in plasma could be disguised by the prevalence of wild-type DNA discharged from normal cells. Secondly, the known heterogeneity of CRC might lead to inconsistency (36). Thirdly, cfDNA in CRC maybe not a sufficiently accurate reflection of tumor biological or clinical status because cfDNA is released mainly from necrotic cells. Whether this discordance is caused by biological or analytical factors remains unclear (37).
Advances in technology are likely to increase the concordance of circulating cfDNA and MPTT DNA genotyping. First of all, improved techniques are required for an optimized extraction of tumor-specific cfDNA. To overcome the limitation of low quantities of cfDNA in the peripheral blood and sequencing artifacts, Newman et al. introduce an approach that integrates in silico elimination of highly stereotypical background artifacts with a molecular barcoding strategy for the further improvement of assay sensitivity (38). Secondly, a hotspot panel that covers the significant mutations related to CRC instead of a large tumor type-agnostic gene panel would help improve the sensitivity and specificity because the amount of cfDNA may be insufficient for whole-exome sequencing (31). Finally, repeated testing in plasma could probably improve the accuracy.
Conlcusions
To summarize, MSI status detection in CRC cfDNA is relatively consistent with those in MPTT samples. Besides, although liquid biopsy is not efficient for tracking tumor mutations of early stage, it may noninvasively provide clues in surveillance of advanced CRC with further optimization. And a larger sample size rigorous verification was needed before specific application of this method in clinical.
Acknowledgments
Funding: The present study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-20-2762
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-20-2762
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-20-2762). All authors report the funding from the National Natural Science Foundation of China, the Guangdong Provincial Natural Science Foundation of China and Guangzhou Science & Technology Project for this study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Nanfang Hospital (Guangzhou, China) (NFEC-2017-193). All patients signed the informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2019;5:1749-68. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Gatalica Z, Vranic S, Xiu J, et al. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer 2016;15:405-12. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol 2013;5:a012633. [Crossref] [PubMed]
- Shia J, Ellis NA, Paty PB, et al. Value of histopathology in predicting microsatellite instability in hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer. Am J Surg Pathol 2003;27:1407-17. [Crossref] [PubMed]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609-18. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov 2015;5:16-8. [Crossref] [PubMed]
- Benson AB, Venook AP, Al-Hawary MM, et al. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. Journal of the National Comprehensive Cancer Network. J Natl Compr Canc Netw 2018;16:359-69. [Crossref] [PubMed]
- Benson AB, Venook AP, Al-Hawary MM, et al. Rectal Cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:874-901. [Crossref] [PubMed]
- Goldstein JB, Wu W, Borras E, et al. Can Microsatellite Status of Colorectal Cancer Be Reliably Assessed after Neoadjuvant Therapy? Clin Cancer Res 2017;23:5246-54. [Crossref] [PubMed]
- Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of Mismatch Repair Deficiency and Microsatellite Instability in Colorectal Adenocarcinoma by Targeted Next-Generation Sequencing. J Mol Diagn 2017;19:84-91. [Crossref] [PubMed]
- Salipante SJ, Scroggins SM, Hampel HL, et al. Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192-9. [Crossref] [PubMed]
- Middha S, Zhang L, Nafa K, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis Oncol 2017;2017:PO.17.00084.
- Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 2018;7:746-56. [Crossref] [PubMed]
- Pietrantonio F, Di Nicolantonio F, Schrock AB, et al. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. J Natl Cancer Inst 2017;109: [Crossref] [PubMed]
- Overman MJ, Lonardi S, Wong KYM, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol 2018;36:773-9. [Crossref] [PubMed]
- Lecomte T, Ceze N, Dorval E, et al. Circulating free tumor DNA and colorectal cancer. Gastroenterol Clin Biol 2010;34:662-81. [Crossref] [PubMed]
- Siravegna G, Bardelli A. Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients. Mol Oncol 2016;10:475-80. [Crossref] [PubMed]
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430-5. [Crossref] [PubMed]
- Lin JK, Lin PC, Lin CH, et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol 2014;21:S680-6. [Crossref] [PubMed]
- Grasselli J, Elez E, Caratu G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 2017;28:1294-301. [Crossref] [PubMed]
- Cuccurullo V, Mansi L. AJCC Cancer Staging Handbook: from the AJCC Cancer Staging Manual (7th edition). Eur J Nucl Med Mol Imaging 2011;38:408.
- Naito S, von Eschenbach AC, Giavazzi R, et al. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res 1986;46:4109-15. [PubMed]
- Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 2015;16:30. [Crossref] [PubMed]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57. [PubMed]
- Meyerhardt JA, Mangu PB, Flynn PJ, et al. Follow-up care, surveillance protocol, and secondary prevention measures for survivors of colorectal cancer: American Society of Clinical Oncology clinical practice guideline endorsement. J Clin Oncol 2013;31:4465-70. [Crossref] [PubMed]
- Willis J, Lefterova MI, Artyomenko A, et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin Cancer Res 2019;25:7035-45. [Crossref] [PubMed]
- Committee of Colorectal Cancer, Chinese Society of Clinical Oncology. Genetics Group of The Committee of Colorectal Cancer, China Anti-Cancer Association; Genetics Committee of The Committee of Colorectal Cancer, Chinese Medical Doctor Association. Consensus on the detection of microsatellite instability in colorectal cancer and other related solid tumors in China. Zhonghua Zhong Liu Za Zhi 2019;41:734-41.
- Furuki H, Yamada T, Takahashi G, et al. Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur J Surg Oncol 2018;44:975-82. [Crossref] [PubMed]
- Barata PC, Koshkin VS, Funchain P, et al. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann Oncol 2017;28:2458-63. [Crossref] [PubMed]
- Jovelet C, Ileana E, Le Deley MC, et al. Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial. Clin Cancer Res 2016;22:2960-8. [Crossref] [PubMed]
- Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715-22. [Crossref] [PubMed]
- Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014;20:4613-24. [Crossref] [PubMed]
- Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 2017;28:1325-32. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]


